Production of biopharmaceutical proteins by yeast: advances through metabolic engineering
- PMID: 23147168
- PMCID: PMC3728191
- DOI: 10.4161/bioe.22856
Production of biopharmaceutical proteins by yeast: advances through metabolic engineering
Abstract
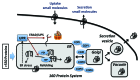
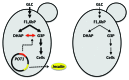
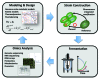
Production of recombinant proteins for use as pharmaceuticals, so-called biopharmaceuticals, is a multi-billion dollar industry. Many different cell factories are used for the production of biopharmaceuticals, but the yeast Saccharomyces cerevisiae is an important cell factory as it is used for production of several large volume products. Insulin and insulin analogs are by far the dominating biopharmaceuticals produced by yeast, and this will increase as the global insulin market is expected to grow from USD12B in 2011 to more than USD32B by 2018. Other important biopharmaceuticals produced by yeast are human serum albumin, hepatitis vaccines and virus like particles used for vaccination against human papillomavirus. Here is given a brief overview of biopharmaceutical production by yeast and it is discussed how the secretory pathway can be engineered to ensure more efficient protein production. The involvement of directed metabolic engineering through the integration of tools from genetic engineering, systems biology and mathematical modeling, is also discussed.
Keywords: Saccharomyces cerevisiae; industrial biotechnology; insulin; secretory pathway; systems biology.
Figures
References
-
- Walsh G. Biopharmaceuticals: Biochemistry and Biotechnology. 1998. J. Wiley & Sons, Ltd., Chichester, UK.
-
- Langer ES. Biomanufacturing outsourcing outlook. BioPharm International. 2012;25:15–6.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
