Kinetic characterization of nonmuscle myosin IIb at the single molecule level
- PMID: 23148220
- PMCID: PMC3537070
- DOI: 10.1074/jbc.M112.424671
Kinetic characterization of nonmuscle myosin IIb at the single molecule level
Abstract
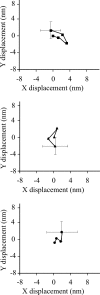
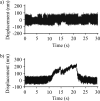
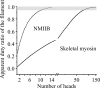
Nonmuscle myosin IIB (NMIIB) is a cytoplasmic myosin, which plays an important role in cell motility by maintaining cortical tension. It forms bipolar thick filaments with ~14 myosin molecule dimers on each side of the bare zone. Our previous studies showed that the NMIIB is a moderately high duty ratio (~20-25%) motor. The ADP release step (~0.35 s(-1)) of NMIIB is only ~3 times faster than the rate-limiting phosphate release (0.13 ± 0.01 s(-1)). The aim of this study was to relate the known in vitro kinetic parameters to the results of single molecule experiments and to compare the kinetic and mechanical properties of single- and double-headed myosin fragments and nonmuscle IIB thick filaments. Examination of the kinetics of NMIIB interaction with actin at the single molecule level was accomplished using total internal reflection fluorescence (TIRF) with fluorescence imaging with 1-nm accuracy (FIONA) and dual-beam optical trapping. At a physiological ATP concentration (1 mm), the rate of detachment of the single-headed and double-headed molecules was similar (~0.4 s(-1)). Using optical tweezers we found that the power stroke sizes of single- and double-headed heavy meromyosin (HMM) were each ~6 nm. No signs of processive stepping at the single molecule level were observed in the case of NMIIB-HMM in optical tweezers or TIRF/in vitro motility experiments. In contrast, robust motility of individual fluorescently labeled thick filaments of full-length NMIIB was observed on actin filaments. Our results are in good agreement with the previous steady-state and transient kinetic studies and show that the individual nonprocessive nonmuscle myosin IIB molecules form a highly processive unit when polymerized into filaments.
Figures




Similar articles
-
Kinetic mechanism of blebbistatin inhibition of nonmuscle myosin IIb.Biochemistry. 2004 Nov 23;43(46):14832-9. doi: 10.1021/bi0490284. Biochemistry. 2004. PMID: 15544354
-
Unconventional processive mechanics of non-muscle myosin IIB.J Biol Chem. 2010 Aug 20;285(34):26326-34. doi: 10.1074/jbc.M110.123851. Epub 2010 May 29. J Biol Chem. 2010. PMID: 20511646 Free PMC article.
-
Myosin-10 produces its power-stroke in two phases and moves processively along a single actin filament under low load.Proc Natl Acad Sci U S A. 2014 May 6;111(18):E1833-42. doi: 10.1073/pnas.1320122111. Epub 2014 Apr 21. Proc Natl Acad Sci U S A. 2014. PMID: 24753602 Free PMC article.
-
Use of fluorescent techniques to study the in vitro movement of myosins.Exp Suppl. 2014;105:193-210. doi: 10.1007/978-3-0348-0856-9_9. Exp Suppl. 2014. PMID: 25095996 Free PMC article. Review.
-
Flexibility in actin-myosin motility system revealed by in vitro motility assay.Adv Biophys. 1991;27:213-20. doi: 10.1016/0065-227x(91)90020-e. Adv Biophys. 1991. PMID: 1755361 Review.
Cited by
-
Unphosphorylated calponin enhances the binding force of unphosphorylated myosin to actin.Biochim Biophys Acta. 2013 Oct;1830(10):4634-41. doi: 10.1016/j.bbagen.2013.05.042. Epub 2013 Jun 6. Biochim Biophys Acta. 2013. PMID: 23747303 Free PMC article.
-
Bond Type and Discretization of Nonmuscle Myosin II Are Critical for Simulated Contractile Dynamics.Biophys J. 2020 Jun 2;118(11):2703-2717. doi: 10.1016/j.bpj.2020.03.033. Epub 2020 Apr 21. Biophys J. 2020. PMID: 32365328 Free PMC article.
-
Myosin II isoform co-assembly and differential regulation in mammalian systems.Exp Cell Res. 2015 May 15;334(1):2-9. doi: 10.1016/j.yexcr.2015.01.012. Epub 2015 Feb 2. Exp Cell Res. 2015. PMID: 25655283 Free PMC article. Review.
-
Myosin 18A coassembles with nonmuscle myosin 2 to form mixed bipolar filaments.Curr Biol. 2015 Mar 30;25(7):942-8. doi: 10.1016/j.cub.2015.02.012. Epub 2015 Mar 5. Curr Biol. 2015. PMID: 25754640 Free PMC article.
-
Kinetic Adaptations of Myosins for Their Diverse Cellular Functions.Traffic. 2016 Aug;17(8):839-59. doi: 10.1111/tra.12388. Epub 2016 Mar 31. Traffic. 2016. PMID: 26929436 Free PMC article. Review.
References
-
- Betapudi V., Licate L. S., Egelhoff T. T. (2006) Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 66, 4725–4733 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

