A direct comparison of next generation sequencing enrichment methods using an aortopathy gene panel- clinical diagnostics perspective
- PMID: 23148498
- PMCID: PMC3534588
- DOI: 10.1186/1755-8794-5-50
A direct comparison of next generation sequencing enrichment methods using an aortopathy gene panel- clinical diagnostics perspective
Abstract
Background: Aortopathies are a group of disorders characterized by aneurysms, dilation, and tortuosity of the aorta. Because of the phenotypic overlap and genetic heterogeneity of diseases featuring aortopathy, molecular testing is often required for timely and correct diagnosis of affected individuals. In this setting next generation sequencing (NGS) offers several advantages over traditional molecular techniques.
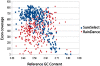
Methods: The purpose of our study was to compare NGS enrichment methods for a clinical assay targeting the nine genes known to be associated with aortopathy. RainDance emulsion PCR and SureSelect RNA-bait hybridization capture enrichment methods were directly compared by enriching DNA from eight samples. Enriched samples were barcoded, pooled, and sequenced on the Illumina HiSeq2000 platform. Depth of coverage, consistency of coverage across samples, and the overlap of variants identified were assessed. This data was also compared to whole-exome sequencing data from ten individuals.
Results: Read depth was greater and less variable among samples that had been enriched using the RNA-bait hybridization capture enrichment method. In addition, samples enriched by hybridization capture had fewer exons with mean coverage less than 10, reducing the need for followup Sanger sequencing. Variants sets produced were 77% concordant, with both techniques yielding similar numbers of discordant variants.
Conclusions: When comparing the design flexibility, performance, and cost of the targeted enrichment methods to whole-exome sequencing, the RNA-bait hybridization capture enrichment gene panel offers the better solution for interrogating the aortopathy genes in a clinical laboratory setting.
Figures



References
-
- Klein DG. Thoracic aortic aneurysms. J Cardiovasc Nurs. 2005;4:245–250. - PubMed
-
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, Xu FL, Myers LA, Spevak PJ, Cameron DE, De Backer J, Hellemans J, Chen Y, Davis EC, Webb CL, Kress W, Coucke P, Rifkin DB, De Paepe AM, Dietz HC. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFΒR1 or TGFΒR2. Nat Genet. 2005;37:275–281. doi: 10.1038/ng1511. - DOI - PubMed
-
- Pannu H, Fadulu VT, Chang J, Lafont A, Hasham SN, Sparks E, Giampietro PF, Zaleski C, Estrera AL, Safi HJ, Shete S, Willing MC, Raman CS, Milewicz DM. Mutations in transforming growth factor-β receptor type II cause familial thoracic aortic aneurysms and dissections. Circulation. 2005;112:513–520. doi: 10.1161/CIRCULATIONAHA.105.537340. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

