A method for high throughput determination of viable bacteria cell counts in 96-well plates
- PMID: 23148795
- PMCID: PMC3534621
- DOI: 10.1186/1471-2180-12-259
A method for high throughput determination of viable bacteria cell counts in 96-well plates
Abstract
Background: There are several methods for quantitating bacterial cells, each with advantages and disadvantages. The most common method is bacterial plating, which has the advantage of allowing live cell assessment through colony forming unit (CFU) counts but is not well suited for high throughput screening (HTS). On the other hand, spectrophotometry is adaptable to HTS applications but does not differentiate between dead and living bacteria and has low sensitivity.
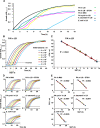
Results: Here, we report a bacterial cell counting method termed Start Growth Time (SGT) that allows rapid and serial quantification of the absolute or relative number of live cells in a bacterial culture in a high throughput manner. We combined the methodology of quantitative polymerase chain reaction (qPCR) calculations with a previously described qualitative method of bacterial growth determination to develop an improved quantitative method. We show that SGT detects only live bacteria and is sensitive enough to differentiate between 40 and 400 cells/mL. SGT is based on the re-growth time required by a growing cell culture to reach a threshold, and the notion that this time is proportional to the number of cells in the initial inoculum. We show several applications of SGT, including assessment of antibiotic effects on cell viability and determination of an antibiotic tolerant subpopulation fraction within a cell population. SGT results do not differ significantly from results obtained by CFU counts.
Conclusion: SGT is a relatively quick, highly sensitive, reproducible and non-laborious method that can be used in HTS settings to longitudinally assess live cells in bacterial cell cultures.
Figures

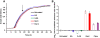
References
-
- Miller JH. In: Experiments in Molecular Genetics. Miller JH, editor. New York: Cold Spring Harbor; 1972. Determination of viable cell counts: bacterial growth curves; pp. 31–36.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

