Photocrosslinking approaches to interactome mapping
- PMID: 23149092
- PMCID: PMC3594551
- DOI: 10.1016/j.cbpa.2012.10.034
Photocrosslinking approaches to interactome mapping
Abstract
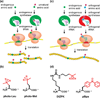
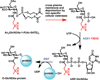
Photocrosslinking approaches can be used to map interactome networks within the context of living cells. Photocrosslinking methods rely on use of metabolic engineering or genetic code expansion to incorporate photocrosslinking analogs of amino acids or sugars into cellular biomolecules. Immunological and mass spectrometry techniques are used to analyze crosslinked complexes, thereby defining specific interactomes. Because photocrosslinking can be conducted in native, cellular settings, it can be used to define context-dependent interactions. Photocrosslinking methods are also ideally suited for determining interactome dynamics, mapping interaction interfaces, and identifying transient interactions in which intrinsically disordered proteins and glycoproteins engage. Here we discuss the application of cell-based photocrosslinking to the study of specific problems in immune cell signaling, transcription, membrane protein dynamics, nucleocytoplasmic transport, and chaperone-assisted protein folding.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



References
-
- Charbonnier S, Gallego O, Gavin AC. The social network of a cell: recent advances in interactome mapping. Biotechnol. Annu. Rev. 2008;14:1–28. - PubMed
-
- Garner AL, Janda KD. Protein-protein interactions and cancer: targeting the central dogma. Curr. Top. Med. Chem. 2011;11:258–280. - PubMed
-
- Tanaka Y, Bond MR, Kohler JJ. Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst. 2008;4:473–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

