Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode
- PMID: 23149449
- PMCID: PMC3565773
- DOI: 10.1038/tp.2012.112
Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode
Abstract
To date, it remains impossible to guarantee that short-term treatment given to a patient suffering from a major depressive episode (MDE) will improve long-term efficacy. Objective biological measurements and biomarkers that could help in predicting the clinical evolution of MDE are still warranted. To better understand the reason nearly half of MDE patients respond poorly to current antidepressive treatments, we examined the gene expression profile of peripheral blood samples collected from 16 severe MDE patients and 13 matched controls. Using a naturalistic and longitudinal design, we ascertained mRNA and microRNA (miRNA) expression at baseline, 2 and 8 weeks later. On a genome-wide scale, we detected transcripts with roles in various biological processes as significantly dysregulated between MDE patients and controls, notably those involved in nucleotide binding and chromatin assembly. We also established putative interactions between dysregulated mRNAs and miRNAs that may contribute to MDE physiopathology. We selected a set of mRNA candidates for quantitative reverse transcriptase PCR (RT-qPCR) to validate that the transcriptional signatures observed in responders is different from nonresponders. Furthermore, we identified a combination of four mRNAs (PPT1, TNF, IL1B and HIST1H1E) that could be predictive of treatment response. Altogether, these results highlight the importance of studies investigating the tight relationship between peripheral transcriptional changes and the dynamic clinical progression of MDE patients to provide biomarkers of MDE evolution and prognosis.
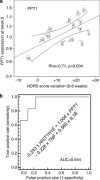
Figures
References
-
- Fekadu A, Wooderson SC, Markopoulo K, Donaldson C, Papadopoulos A, Cleare AJ. What happens to patients with treatment-resistant depression? A systematic review of medium to long term outcome studies. J Affect Disord. 2009;116:4–11. - PubMed
-
- American Psychiatric Association Practice Guideline for the Treatment of Patients with Major Depressive Disorder3rd edn.American Psychiatric Association (APA): Arlington, VA; 2010152
-
- Kennedy SH, Lam RW, Parikh SV, Patten SB, Ravindran AV. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults. Introduction. J Affect Disord. 2009;117 (Suppl 1:S1–S2. - PubMed
-
- Bauer M, Whybrow PC, Angst J, Versiani M, Moller HJ. World Federation of Societies of Biological Psychiatry (WFSBP) Guidelines for Biological Treatment of Unipolar Depressive Disorders, Part 1: Acute and continuation treatment of major depressive disorder. World J Biol Psychiatry. 2002;3:5–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

