RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues
- PMID: 23149937
- PMCID: PMC3554129
- DOI: 10.1128/MCB.01174-12
RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues
Abstract
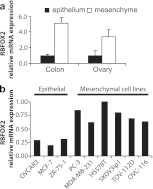
Alternative splicing provides a critical and flexible layer of regulation intervening in many biological processes to regulate the diversity of proteins and impact cell phenotype. To identify alternative splicing differences that distinguish epithelial from mesenchymal tissues, we have investigated hundreds of cassette exons using a high-throughput reverse transcription-PCR (RT-PCR) platform. Extensive changes in splicing were noted between epithelial and mesenchymal tissues in both human colon and ovarian tissues, with many changes from mostly one splice variant to predominantly the other. Remarkably, many of the splicing differences that distinguish normal mesenchymal from normal epithelial tissues matched those that differentiate normal ovarian tissues from ovarian cancer. Furthermore, because splicing profiling could classify cancer cell lines according to their epithelial/mesenchymal characteristics, we used these cancer cell lines to identify regulators for these specific splicing signatures. By knocking down 78 potential splicing factors in five cell lines, we provide an extensive view of the complex regulatory landscape associated with the epithelial and mesenchymal states, thus revealing that RBFOX2 is an important driver of mesenchymal tissue-specific splicing.
Figures




References
-
- Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890 - PubMed
-
- Barrallo-Gimeno A, Nieto MA. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132:3151–3161 - PubMed
-
- Polyak K, Weinberg RA. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9:265–273 - PubMed
-
- Yilmaz M, Christofori G. 2009. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 28:15–33 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
