The role of DNA methylation and histone modifications in transcriptional regulation in humans
- PMID: 23150256
- PMCID: PMC6611551
- DOI: 10.1007/978-94-007-4525-4_13
The role of DNA methylation and histone modifications in transcriptional regulation in humans
Abstract
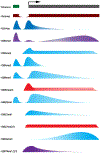
Although the field of genetics has grown by leaps and bounds within the last decade due to the completion and availability of the human genome sequence, transcriptional regulation still cannot be explained solely by an individual's DNA sequence. Complex coordination and communication between a plethora of well-conserved chromatin modifying factors are essential for all organisms. Regulation of gene expression depends on histone post translational modifications (HPTMs), DNA methylation, histone variants, remodeling enzymes, and effector proteins that influence the structure and function of chromatin, which affects a broad spectrum of cellular processes such as DNA repair, DNA replication, growth, and proliferation. If mutated or deleted, many of these factors can result in human disease at the level of transcriptional regulation. The common goal of recent studies is to understand disease states at the stage of altered gene expression. Utilizing information gained from new high-throughput techniques and analyses will aid biomedical research in the development of treatments that work at one of the most basic levels of gene expression, chromatin. This chapter will discuss the effects of and mechanism by which histone modifications and DNA methylation affect transcriptional regulation.
Figures
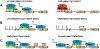
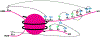
References
-
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature, 449(7163), 731–734. - PubMed
-
- Allis CD, Bowen JK, Abraham GN, Glover CV, & Gorovsky MA (1980). Proteolytic processing of histone H3 in chromatin: A physiologically regulated event in tetrahymena micronuclei. Cell, 20(1), 55–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

