Bioinspired multivalent DNA network for capture and release of cells
- PMID: 23150586
- PMCID: PMC3511714
- DOI: 10.1073/pnas.1211234109
Bioinspired multivalent DNA network for capture and release of cells
Abstract
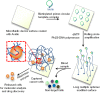
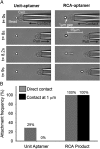
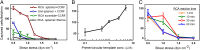
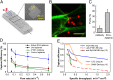
Capture and isolation of flowing cells and particulates from body fluids has enormous implications in diagnosis, monitoring, and drug testing, yet monovalent adhesion molecules used for this purpose result in inefficient cell capture and difficulty in retrieving the captured cells. Inspired by marine creatures that present long tentacles containing multiple adhesive domains to effectively capture flowing food particulates, we developed a platform approach to capture and isolate cells using a 3D DNA network comprising repeating adhesive aptamer domains that extend over tens of micrometers into the solution. The DNA network was synthesized from a microfluidic surface by rolling circle amplification where critical parameters, including DNA graft density, length, and sequence, could readily be tailored. Using an aptamer that binds to protein tyrosine kinase-7 (PTK7) that is overexpressed on many human cancer cells, we demonstrate that the 3D DNA network significantly enhances the capture efficiency of lymphoblast CCRF-CEM cells over monovalent aptamers and antibodies, yet maintains a high purity of the captured cells. When incorporated in a herringbone microfluidic device, the 3D DNA network not only possessed significantly higher capture efficiency than monovalent aptamers and antibodies, but also outperformed previously reported cell-capture microfluidic devices at high flow rates. This work suggests that 3D DNA networks may have broad implications for detection and isolation of cells and other bioparticles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8(5):329–340. - PubMed
-
- Dharmasiri U, Witek MA, Adams AA, Soper SA. Microsystems for the capture of low-abundance cells. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:409–431. - PubMed
-
- Riethdorf S, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

