Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana
- PMID: 23150599
- PMCID: PMC3527241
- DOI: 10.1534/genetics.112.146100
Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana
Abstract
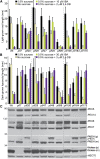
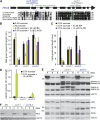
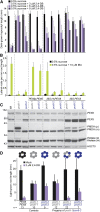
Peroxisomes are organelles that sequester certain metabolic pathways; many of these pathways generate H(2)O(2), which can damage proteins. However, little is known about how damaged or obsolete peroxisomal proteins are degraded. We exploit developmentally timed peroxisomal content remodeling in Arabidopsis thaliana to elucidate peroxisome-associated protein degradation. Isocitrate lyase (ICL) is a peroxisomal glyoxylate cycle enzyme necessary for early seedling development. A few days after germination, photosynthesis begins and ICL is degraded. We previously found that ICL is stabilized when a peroxisome-associated ubiquitin-conjugating enzyme and its membrane anchor are both mutated, suggesting that matrix proteins might exit the peroxisome for ubiquitin-dependent cytosolic degradation. To identify additional components needed for peroxisome-associated matrix protein degradation, we mutagenized a line expressing GFP-ICL, which is degraded similarly to endogenous ICL, and identified persistent GFP-ICL fluorescence (pfl) mutants. We found three pfl mutants that were defective in PEROXIN14 (PEX14/At5g62810), which encodes a peroxisomal membrane protein that assists in importing proteins into the peroxisome matrix, indicating that proteins must enter the peroxisome for efficient degradation. One pfl mutant was missing the peroxisomal 3-ketoacyl-CoA thiolase encoded by the PEROXISOME DEFECTIVE1 (PED1/At2g33150) gene, suggesting that peroxisomal metabolism influences the rate of matrix protein degradation. Finally, one pfl mutant that displayed normal matrix protein import carried a novel lesion in PEROXIN6 (PEX6/At1g03000), which encodes a peroxisome-tethered ATPase that is involved in recycling matrix protein receptors back to the cytosol. The isolation of pex6-2 as a pfl mutant supports the hypothesis that matrix proteins can exit the peroxisome for cytosolic degradation.
Figures





References
-
- Adham A. R., Zolman B. K., Millius A., Bartel B., 2005. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J. 41: 859–874 - PubMed
-
- Azevedo J. E., Schliebs W., 2006. Pex14p, more than just a docking protein. Biochim. Biophys. Acta 1763: 1574–1584 - PubMed
-
- Bell C. J., Ecker J. R., 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

