Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution
- PMID: 23150601
- PMCID: PMC3567732
- DOI: 10.1534/genetics.112.145631
Long-range targeted manipulation of the Drosophila genome by site-specific integration and recombinational resolution
Abstract
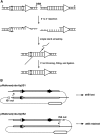
Significant advances in genomics underscore the importance of targeted mutagenesis for gene function analysis. Here we have developed a scheme for long-range targeted manipulation of genes in the Drosophila genome. Utilizing an attP attachment site for the phiC31 integrase previously targeted to the nbs gene, we integrated an 80-kb genomic fragment at its endogenous locus to generate a tandem duplication of the region. We achieved reduction to a single copy by inducing recombination via a site-specific DNA break. We report that, despite the large size of the DNA fragment, both plasmid integration and duplication reduction can be accomplished efficiently. Importantly, the integrating genomic fragment can serve as a venue for introducing targeted modifications to the entire region. We successfully introduced a new attachment site 70 kb from the existing attP using this two-step scheme, making a new region susceptible to targeted mutagenesis. By experimenting with different placements of the future DNA break site in the integrating vector, we established a vector configuration that facilitates the recovery of desired modifications. We also show that reduction events can occur efficiently through unequal meiotic crossing over between the large duplications. Based on our results, we suggest that a collection of 1200 lines with attachment sites inserted every 140 kb throughout the genome would render all Drosophila genes amenable to targeted mutagenesis. Excitingly, all of the components involved are likely functional in other eukaryotes, making our scheme for long-range targeted manipulation readily applicable to other systems.
Figures



Comment in
-
Targeted gene replacement in Drosophila goes the distance.Genetics. 2013 Feb;193(2):377-81. doi: 10.1534/genetics.112.148379. Genetics. 2013. PMID: 23396476 Free PMC article. No abstract available.
References
-
- Copeland N. G., Jenkins N. A., Court D. L., 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2: 769–779 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

