Differential network entropy reveals cancer system hallmarks
- PMID: 23150773
- PMCID: PMC3496163
- DOI: 10.1038/srep00802
Differential network entropy reveals cancer system hallmarks
Abstract
The cellular phenotype is described by a complex network of molecular interactions. Elucidating network properties that distinguish disease from the healthy cellular state is therefore of critical importance for gaining systems-level insights into disease mechanisms and ultimately for developing improved therapies. By integrating gene expression data with a protein interaction network we here demonstrate that cancer cells are characterised by an increase in network entropy. In addition, we formally demonstrate that gene expression differences between normal and cancer tissue are anticorrelated with local network entropy changes, thus providing a systemic link between gene expression changes at the nodes and their local correlation patterns. In particular, we find that genes which drive cell-proliferation in cancer cells and which often encode oncogenes are associated with reductions in network entropy. These findings may have potential implications for identifying novel drug targets.
Figures

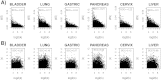
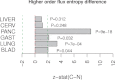
 measure (equation 2) which is defined for a stochastic diffusion matrix for maximum path lengths of order 2 (Methods). Positive z-statistics means higher entropy in cancer compared to normal. Green lines indicate the 95% confidence interval envelope and given P-values are from a normal null distribution centred at zero.
measure (equation 2) which is defined for a stochastic diffusion matrix for maximum path lengths of order 2 (Methods). Positive z-statistics means higher entropy in cancer compared to normal. Green lines indicate the 95% confidence interval envelope and given P-values are from a normal null distribution centred at zero.References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

