Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini
- PMID: 23151128
- PMCID: PMC3546061
- DOI: 10.1186/1471-2164-13-622
Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini
Abstract
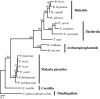
Background: Mitochondrial (mt) genomes vary considerably in size, structure and gene content. The mt genomes of the phylum Apicomplexa, which includes important human pathogens such as the malaria parasite Plasmodium, also show marked diversity of structure. Plasmodium has a concatenated linear mt genome of the smallest size (6-kb); Babesia and Theileria have a linear monomeric mt genome (6.5-kb to 8.2-kb) with terminal inverted repeats; Eimeria, which is distantly related to Plasmodium and Babesia/Theileria, possesses a mt genome (6.2-kb) with a concatemeric form similar to that of Plasmodium; Cryptosporidium, the earliest branching lineage within the phylum Apicomplexa, has no mt genome. We are interested in the evolutionary origin of linear mt genomes of Babesia/Theileria, and have investigated mt genome structures in members of archaeopiroplasmid, a lineage branched off earlier from Babesia/Theileria.
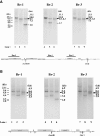
Results: The complete mt genomes of archaeopiroplasmid parasites, Babesia microti and Babesia rodhaini, were sequenced. The mt genomes of B. microti (11.1-kb) and B. rodhaini (6.9-kb) possess two pairs of unique inverted repeats, IR-A and IR-B. Flip-flop inversions between two IR-As and between two IR-Bs appear to generate four distinct genome structures that are present at an equi-molar ratio. An individual parasite contained multiple mt genome structures, with 20 copies and 2 - 3 copies per haploid nuclear genome in B. microti and B. rodhaini, respectively.
Conclusion: We found a novel linear monomeric mt genome structure of B. microti and B. rhodhaini equipped with dual flip-flop inversion system, by which four distinct genome structures are readily generated. To our knowledge, this study is the first to report the presence of two pairs of distinct IR sequences within a monomeric linear mt genome. The present finding provides insight into further understanding of evolution of mt genome structure.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

