The size and conservation of a coiled-coil structure in the ectodomain of human BST-2/tetherin is dispensable for inhibition of HIV-1 virion release
- PMID: 23152502
- PMCID: PMC3531743
- DOI: 10.1074/jbc.M112.418822
The size and conservation of a coiled-coil structure in the ectodomain of human BST-2/tetherin is dispensable for inhibition of HIV-1 virion release
Abstract
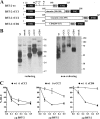
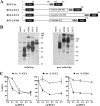
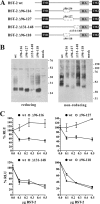
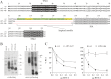
BST-2/CD317/tetherin is a host factor that inhibits HIV-1 release and is counteracted by HIV-1 Vpu. Structural studies indicate that the BST-2 ectodomain assumes a coiled-coil conformation. Here we studied the role of the BST-2 ectodomain for tethering function. First, we addressed the importance of the length and structure of the ectodomain by adding or substituting heterologous coiled-coil or non-coiled-coil sequences. We found that extending or replacing the BST-2 ectodomain using non-coiled-coil sequences resulted in loss of BST-2 function. Doubling the size of the BST-2 ectodomain by insertion of a heterologous coiled-coil motif or substituting the BST-2 coiled-coil domain with a heterologous coiled-coil motif maintained tethering function. Reductions in the size of the BST-2 coiled-coil domain were tolerated as well. In fact, deletion of the C-terminal half of the BST-2 ectodomain, including a series of seven consecutive heptad motifs did not abolish tethering function. However, slight changes in the positioning of deletions affecting the relative placing of charged or hydrophobic residues on the helix severely impacted the functional properties of BST-2. Overall, we conclude that the size of the BST-2 ectodomain is highly flexible and can be reduced or extended as long as the positioning of residues important for the stability of the dimer interface is maintained.
Figures

Similar articles
-
The tetherin/BST-2 coiled-coil ectodomain mediates plasma membrane microdomain localization and restriction of particle release.J Virol. 2012 Feb;86(4):2259-72. doi: 10.1128/JVI.05906-11. Epub 2011 Nov 30. J Virol. 2012. PMID: 22130541 Free PMC article.
-
Positioning of cysteine residues within the N-terminal portion of the BST-2/tetherin ectodomain is important for functional dimerization of BST-2.J Biol Chem. 2015 Feb 6;290(6):3740-51. doi: 10.1074/jbc.M114.617639. Epub 2014 Dec 18. J Biol Chem. 2015. PMID: 25525265 Free PMC article.
-
[Inhibition of HIV virus-like particles production by BST-2].Bing Du Xue Bao. 2011 Jul;27(4):319-25. Bing Du Xue Bao. 2011. PMID: 21874899 Chinese.
-
Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction.Viruses. 2011 May;3(5):520-40. doi: 10.3390/v3050520. Epub 2011 May 6. Viruses. 2011. PMID: 21994744 Free PMC article. Review.
-
Tetherin/BST-2: Restriction Factor or Immunomodulator?Curr HIV Res. 2016;14(3):235-46. doi: 10.2174/1570162x14999160224102752. Curr HIV Res. 2016. PMID: 26957198 Free PMC article. Review.
Cited by
-
Multi-functional BST2/tetherin against HIV-1, other viruses and LINE-1.Front Cell Infect Microbiol. 2022 Sep 13;12:979091. doi: 10.3389/fcimb.2022.979091. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36176574 Free PMC article. Review.
-
Resilience of BST-2/Tetherin structure to single amino acid substitutions.PeerJ. 2019 May 31;7:e7043. doi: 10.7717/peerj.7043. eCollection 2019. PeerJ. 2019. PMID: 31183261 Free PMC article.
-
Functional antagonism of rhesus macaque and chimpanzee BST-2 by HIV-1 Vpu is mediated by cytoplasmic domain interactions.J Virol. 2013 Dec;87(24):13825-36. doi: 10.1128/JVI.02567-13. Epub 2013 Oct 9. J Virol. 2013. PMID: 24109238 Free PMC article.
-
HIV-1 Vpu - an ion channel in search of a job.Biochim Biophys Acta. 2014 Apr;1838(4):1074-81. doi: 10.1016/j.bbamem.2013.06.029. Epub 2013 Jul 3. Biochim Biophys Acta. 2014. PMID: 23831603 Free PMC article. Review.
-
HIV accessory proteins versus host restriction factors.Curr Opin Virol. 2013 Dec;3(6):692-9. doi: 10.1016/j.coviro.2013.08.004. Epub 2013 Nov 15. Curr Opin Virol. 2013. PMID: 24246762 Free PMC article. Review.
References
-
- Neil S. J., Zang T., Bieniasz P. D. (2008) Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

