Inhibition of 5'-UTR RNA conformational switching in HIV-1 using antisense PNAs
- PMID: 23152893
- PMCID: PMC3495914
- DOI: 10.1371/journal.pone.0049310
Inhibition of 5'-UTR RNA conformational switching in HIV-1 using antisense PNAs
Abstract
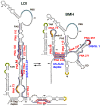
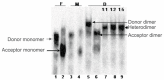
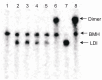
Background: The genome of retroviruses, including HIV-1, is packaged as two homologous (+) strand RNA molecules, noncovalently associated close to their 5'-end in a region called dimer linkage structure (DLS). Retroviral HIV-1 genomic RNAs dimerize through complex interactions between dimerization initiation sites (DIS) within the (5'-UTR). Dimer formation is prevented by so calledLong Distance Interaction (LDI) conformation, whereas Branched Multiple Hairpin (BMH) conformation leads to spontaneous dimerization.
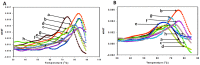
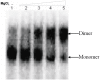
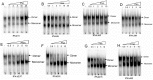
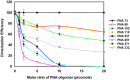
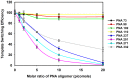
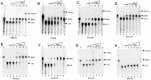
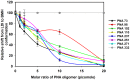
Methods and results: We evaluated the role of SL1 (DIS), PolyA Hairpin signal and a long distance U5-AUG interaction by in-vitro dimerization, conformer assay and coupled dimerization and template-switching assays using antisense PNAs. Our data suggests evidence that PNAs targeted against SL1 produced severe inhibitory effect on dimerization and template-switching processes while PNAs targeted against U5 region do not show significant effect on dimerization and template switching, while PNAs targeted against AUG region showed strong inhibition of dimerization and template switching processes.
Conclusions: Our results demonstrate that PNA can be used successfully as an antisense to inhibit dimerization and template switching process in HIV -1 and both of the processes are closely linked to each other. Different PNA oligomers have ability of switching between two thermodynamically stable forms. PNA targeted against DIS and SL1 switch, LDI conformer to more dimerization friendly BMH form. PNAs targeted against PolyA haipin configuration did not show a significant change in dimerization and template switching process. The PNA oligomer directed against the AUG strand of U5-AUG duplex structure also showed a significant reduction in RNA dimerization as well as template- switching efficiency.The antisense PNA oligomers can be used to regulate the shift in the LDI/BMH equilibrium.
Conflict of interest statement
Figures


Similar articles
-
The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation.Biochemistry. 2005 Jun 28;44(25):9058-66. doi: 10.1021/bi0502588. Biochemistry. 2005. PMID: 15966729
-
Role of the 5' TAR stem--loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA.Biochemistry. 2008 Mar 11;47(10):3283-93. doi: 10.1021/bi7023173. Epub 2008 Feb 16. Biochemistry. 2008. PMID: 18278873
-
In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes.J Biol Chem. 2002 May 31;277(22):19967-75. doi: 10.1074/jbc.M200950200. Epub 2002 Mar 14. J Biol Chem. 2002. PMID: 11896057
-
Dimerization of retroviral genomic RNAs: structural and functional implications.Biochimie. 1996;78(7):639-53. doi: 10.1016/s0300-9084(96)80010-1. Biochimie. 1996. PMID: 8955907 Review.
-
Peptide nucleic acids (PNAs) antisense effect to bacterial growth and their application potentiality in biotechnology.Appl Microbiol Biotechnol. 2010 Mar;86(2):397-402. doi: 10.1007/s00253-009-2387-8. Epub 2010 Feb 5. Appl Microbiol Biotechnol. 2010. PMID: 20135118 Review.
Cited by
-
Machine Learning Guides Peptide Nucleic Acid Flow Synthesis and Sequence Design.Adv Sci (Weinh). 2022 Dec;9(34):e2201988. doi: 10.1002/advs.202201988. Epub 2022 Oct 21. Adv Sci (Weinh). 2022. PMID: 36270977 Free PMC article.
-
Dual Mechanisms of Action of Self-Delivering, Anti-HIV-1 FANA Oligonucleotides as a Potential New Approach to HIV Therapy.Mol Ther Nucleic Acids. 2019 Sep 6;17:615-625. doi: 10.1016/j.omtn.2019.07.001. Epub 2019 Jul 11. Mol Ther Nucleic Acids. 2019. PMID: 31394430 Free PMC article.
-
Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging.RNA. 2013 Aug;19(8):1078-88. doi: 10.1261/rna.038869.113. Epub 2013 Jun 24. RNA. 2013. PMID: 23798665 Free PMC article.
-
Synthesis of peptide nucleic acids containing a crosslinking agent and evaluation of their reactivities.Molecules. 2015 Mar 13;20(3):4708-19. doi: 10.3390/molecules20034708. Molecules. 2015. PMID: 25781072 Free PMC article.
References
-
- Bender W, Davidson N (1976) Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7: 595–607. - PubMed
-
- Hoglund S, Ohagen A, Goncalves J, Panganiban AT, Gabuzda D (1997) Ultrastructure of HIV-1 genomic RNA. Virology 233: 271–279. - PubMed
-
- Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barre-Sinoussi F (1990) Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol 216: 689–699. - PubMed
-
- Gotte M, Li X, Wainberg MA (1999) HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch Biochem Biophys 365: 199–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

