Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation
- PMID: 23153532
- PMCID: PMC3512565
- DOI: 10.1016/j.ccr.2012.08.013
Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation
Abstract
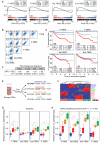
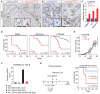
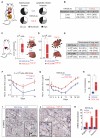
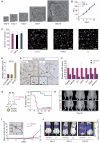
A large proportion of colorectal cancers (CRCs) display mutational inactivation of the TGF-β pathway, yet, paradoxically, they are characterized by elevated TGF-β production. Here, we unveil a prometastatic program induced by TGF-β in the microenvironment that associates with a high risk of CRC relapse upon treatment. The activity of TGF-β on stromal cells increases the efficiency of organ colonization by CRC cells, whereas mice treated with a pharmacological inhibitor of TGFBR1 are resilient to metastasis formation. Secretion of IL11 by TGF-β-stimulated cancer-associated fibroblasts (CAFs) triggers GP130/STAT3 signaling in tumor cells. This crosstalk confers a survival advantage to metastatic cells. The dependency on the TGF-β stromal program for metastasis initiation could be exploited to improve the diagnosis and treatment of CRC.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Metastatic ability: adapting to a tissue site unseen.Cancer Cell. 2012 Nov 13;22(5):563-4. doi: 10.1016/j.ccr.2012.10.011. Cancer Cell. 2012. PMID: 23153528 Free PMC article.
References
-
- Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, Nikolaev A, Burg J, Strand S, Kiesslich R, Huber S, et al. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21:491–501. - PubMed
-
- Bhatia M, Davenport V, Cairo MS. The role of interleukin-11 to prevent chemotherapy-induced thrombocytopenia in patients with solid tumors, lymphoma, acute myeloid leukemia and bone marrow failure syndromes. Leuk Lymphoma. 2007;48:9–15. - PubMed
-
- Bueno L, de Alwis DP, Pitou C, Yingling J, Lahn M, Glatt S, Troconiz IF. Semi-mechanistic modelling of the tumour growth inhibitory effects of LY2157299, a new type I receptor TGF-beta kinase antagonist, in mice. Eur J Cancer. 2008;44:142–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

