Dysregulation of RNA polymerase I transcription during disease
- PMID: 23153826
- PMCID: PMC3594452
- DOI: 10.1016/j.bbagrm.2012.10.014
Dysregulation of RNA polymerase I transcription during disease
Abstract
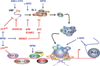
Transcription of the ribosomal RNA genes by the dedicated RNA polymerase I enzyme and subsequent processing of the ribosomal RNA are fundamental control steps in the synthesis of functional ribosomes. Dysregulation of Pol I transcription and ribosome biogenesis is linked to the etiology of a broad range of human diseases. Diseases caused by loss of function mutations in the molecular constituents of the ribosome, or factors intimately associated with RNA polymerase I transcription and processing are collectively termed ribosomopathies. Ribosomopathies are generally rare and treatment options are extremely limited tending to be more palliative than curative. Other more common diseases are associated with profound changes in cellular growth such as cardiac hypertrophy, atrophy or cancer. In contrast to ribosomopathies, altered RNA polymerase I transcriptional activity in these diseases largely results from dysregulated upstream oncogenic pathways or by direct modulation by oncogenes or tumor suppressors at the level of the RNA polymerase I transcription apparatus itself. Ribosomopathies associated with mutations in ribosomal proteins and ribosomal RNA processing or assembly factors have been covered by recent excellent reviews. In contrast, here we review our current knowledge of human diseases specifically associated with dysregulation of RNA polymerase I transcription and its associated regulatory apparatus, including some cases where this dysregulation is directly causative in disease. We will also provide insight into and discussion of possible therapeutic approaches to treat patients with dysregulated RNA polymerase I transcription. This article is part of a Special Issue entitled: Transcription by Odd Pols.
Copyright © 2012 Elsevier B.V. All rights reserved.
Conflict of interest statement
R.D. Hannan is a member of the Scientific Advisory Board for Cylene.
Figures


References
-
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. - PubMed
-
- Grummt I. Wisely chosen paths--regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden. FEBS J. 2010;277:4626–4639. - PubMed
-
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. - PubMed
-
- Narla A, Ebert BL. Translational medicine: ribosomopathies. Blood. 2011;118:4300–4301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

