Recognition of CD1d-restricted antigens by natural killer T cells
- PMID: 23154222
- PMCID: PMC3740582
- DOI: 10.1038/nri3328
Recognition of CD1d-restricted antigens by natural killer T cells
Abstract
Natural killer T (NKT) cells are innate-like T cells that rapidly produce a variety of cytokines following T cell receptor (TCR) activation and can shape the immune response in many different settings. There are two main NKT cell subsets: type I NKT cells are typically characterized by the expression of a semi-invariant TCR, whereas the TCRs expressed by type II NKT cells are more diverse. This Review focuses on the defining features and emerging generalities regarding how NKT cells specifically recognize self, microbial and synthetic lipid-based antigens that are presented by CD1d. Such information is vitally important to better understand, and fully harness, the therapeutic potential of NKT cells.
Conflict of interest statement
The authors declare no competing financial interests.
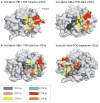
Figures





References
-
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. - PubMed
-
- Mori L, De Libero G. T cells specific for lipid antigens. Immunol Res. 2012;53:191–199. - PubMed
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
-
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nature Rev Immunol. 2004;4:231–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

