Nucleoside triphosphates--building blocks for the modification of nucleic acids
- PMID: 23154273
- PMCID: PMC6268876
- DOI: 10.3390/molecules171113569
Nucleoside triphosphates--building blocks for the modification of nucleic acids
Abstract
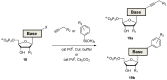
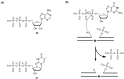
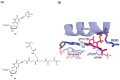
Nucleoside triphosphates are moldable entities that can easily be functionalized at various locations. The enzymatic polymerization of these modified triphosphate analogues represents a versatile platform for the facile and mild generation of (highly) functionalized nucleic acids. Numerous modified triphosphates have been utilized in a broad palette of applications spanning from DNA-tagging and -labeling to the generation of catalytic nucleic acids. This review will focus on the recent progress made in the synthesis of modified nucleoside triphosphates as well as on the understanding of the mechanisms underlying their polymerase acceptance. In addition, the usefulness of chemically altered dNTPs in SELEX and related methods of in vitro selection will be highlighted, with a particular emphasis on the generation of modified DNA enzymes (DNAzymes) and DNA-based aptamers.
Figures
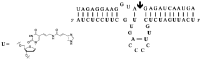

Similar articles
-
The Toolbox for Modified Aptamers.Mol Biotechnol. 2016 Feb;58(2):79-92. doi: 10.1007/s12033-015-9907-9. Mol Biotechnol. 2016. PMID: 26607475 Review.
-
Nucleoside triphosphates--from synthesis to biochemical characterization.J Vis Exp. 2014 Apr 3;(86):51385. doi: 10.3791/51385. J Vis Exp. 2014. PMID: 24747811 Free PMC article.
-
Expanding the catalytic repertoire of DNAzymes by modified nucleosides.Chimia (Aarau). 2011;65(10):770-5. doi: 10.2533/chimia.2011.770. Chimia (Aarau). 2011. PMID: 22054129
-
Enzymatic and chemical synthesis of isodeoxynucleic acid (INA) and its properties.Nucleic Acids Symp Ser (Oxf). 2007;(51):355-6. doi: 10.1093/nass/nrm178. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029733
-
Enzymatic recognition of 2'-modified ribonucleoside 5'-triphosphates: towards the evolution of versatile aptamers.Chembiochem. 2012 Jan 2;13(1):19-25. doi: 10.1002/cbic.201100648. Epub 2011 Dec 12. Chembiochem. 2012. PMID: 22162282 Review.
Cited by
-
Synthesis and Biological Profiling of Quinolino-Fused 7-Deazapurine Nucleosides.ACS Omega. 2024 Apr 27;9(18):20557-20570. doi: 10.1021/acsomega.4c02031. eCollection 2024 May 7. ACS Omega. 2024. PMID: 38737052 Free PMC article.
-
Generation of Aptamers with an Expanded Chemical Repertoire.Molecules. 2015 Sep 14;20(9):16643-71. doi: 10.3390/molecules200916643. Molecules. 2015. PMID: 26389865 Free PMC article. Review.
-
Efficient enzymatic synthesis and dual-colour fluorescent labelling of DNA probes using long chain azido-dUTP and BCN dyes.Nucleic Acids Res. 2016 May 5;44(8):e79. doi: 10.1093/nar/gkw028. Epub 2016 Jan 26. Nucleic Acids Res. 2016. PMID: 26819406 Free PMC article.
-
Modified Nucleoside Triphosphates for In-vitro Selection Techniques.Front Chem. 2016 May 4;4:18. doi: 10.3389/fchem.2016.00018. eCollection 2016. Front Chem. 2016. PMID: 27200340 Free PMC article. Review.
-
Tethered imidazole mediated duplex stabilization and its potential for aptamer stabilization.Nucleic Acids Res. 2018 Dec 14;46(22):11671-11686. doi: 10.1093/nar/gky1062. Nucleic Acids Res. 2018. PMID: 30418582 Free PMC article.
References
-
- Tuerk C., Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

