Ancient properties of spider silks revealed by the complete gene sequence of the prey-wrapping silk protein (AcSp1)
- PMID: 23155003
- PMCID: PMC3563967
- DOI: 10.1093/molbev/mss254
Ancient properties of spider silks revealed by the complete gene sequence of the prey-wrapping silk protein (AcSp1)
Abstract
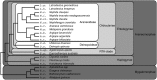
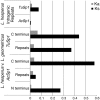
Spider silk fibers have impressive mechanical properties and are primarily composed of highly repetitive structural proteins (termed spidroins) encoded by a single gene family. Most characterized spidroin genes are incompletely known because of their extreme size (typically >9 kb) and repetitiveness, limiting understanding of the evolutionary processes that gave rise to their unusual gene architectures. The only complete spidroin genes characterized thus far form the dragline in the Western black widow, Latrodectus hesperus. Here, we describe the first complete gene sequence encoding the aciniform spidroin AcSp1, the primary component of spider prey-wrapping fibers. L. hesperus AcSp1 contains a single enormous (∼19 kb) exon. The AcSp1 repeat sequence is exceptionally conserved between two widow species (∼94% identity) and between widows and distantly related orb-weavers (∼30% identity), consistent with a history of strong purifying selection on its amino acid sequence. Furthermore, the 16 repeats (each 371-375 amino acids long) found in black widow AcSp1 are, on average, >99% identical at the nucleotide level. A combination of stabilizing selection on amino acid sequence, selection on silent sites, and intragenic recombination likely explains the extreme homogenization of AcSp1 repeats. In addition, phylogenetic analyses of spidroin paralogs support a gene duplication event occurring concomitantly with specialization of the aciniform glands and the tubuliform glands, which synthesize egg-case silk. With repeats that are dramatically different in length and amino acid composition from dragline spidroins, our L. hesperus AcSp1 expands the knowledge base for developing silk-based biomimetic technologies.
Figures



References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465:236–238. - PubMed
-
- Ayoub NA, Hayashi CY. Multiple recombining loci encode MaSp1, the primary constituent of dragline silk, in widow spiders (Latrodectus: Theridiidae) Mol Biol Evol. 2008;25:277–286. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

