Novel β-catenin target genes identified in thalamic neurons encode modulators of neuronal excitability
- PMID: 23157480
- PMCID: PMC3532193
- DOI: 10.1186/1471-2164-13-635
Novel β-catenin target genes identified in thalamic neurons encode modulators of neuronal excitability
Abstract
Background: LEF1/TCF transcription factors and their activator β-catenin are effectors of the canonical Wnt pathway. Although Wnt/β-catenin signaling has been implicated in neurodegenerative and psychiatric disorders, its possible role in the adult brain remains enigmatic. To address this issue, we sought to identify the genetic program activated by β-catenin in neurons. We recently showed that β-catenin accumulates specifically in thalamic neurons where it activates Cacna1g gene expression. In the present study, we combined bioinformatics and experimental approaches to find new β-catenin targets in the adult thalamus.
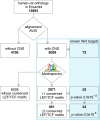
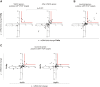
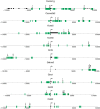
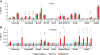
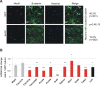
Results: We first selected the genes with at least two conserved LEF/TCF motifs within the regulatory elements. The resulting list of 428 putative LEF1/TCF targets was significantly enriched in known Wnt targets, validating our approach. Functional annotation of the presumed targets also revealed a group of 41 genes, heretofore not associated with Wnt pathway activity, that encode proteins involved in neuronal signal transmission. Using custom polymerase chain reaction arrays, we profiled the expression of these genes in the rat forebrain. We found that nine of the analyzed genes were highly expressed in the thalamus compared with the cortex and hippocampus. Removal of nuclear β-catenin from thalamic neurons in vitro by introducing its negative regulator Axin2 reduced the expression of six of the nine genes. Immunoprecipitation of chromatin from the brain tissues confirmed the interaction between β-catenin and some of the predicted LEF1/TCF motifs. The results of these experiments validated four genes as authentic and direct targets of β-catenin: Gabra3 for the receptor of GABA neurotransmitter, Calb2 for the Ca(2+)-binding protein calretinin, and the Cacna1g and Kcna6 genes for voltage-gated ion channels. Two other genes from the latter cluster, Cacna2d2 and Kcnh8, appeared to be regulated by β-catenin, although the binding of β-catenin to the regulatory sequences of these genes could not be confirmed.
Conclusions: In the thalamus, β-catenin regulates the expression of a novel group of genes that encode proteins involved in neuronal excitation. This implies that the transcriptional activity of β-catenin is necessary for the proper excitability of thalamic neurons, may influence activity in the thalamocortical circuit, and may contribute to thalamic pathologies.
Figures

References
-
- Brembeck FH, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16(1):51–59. - PubMed
-
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. - PubMed
-
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

