Anti-HBV efficacy of combined siRNAs targeting viral gene and heat shock cognate 70
- PMID: 23158906
- PMCID: PMC3534549
- DOI: 10.1186/1743-422X-9-275
Anti-HBV efficacy of combined siRNAs targeting viral gene and heat shock cognate 70
Abstract
Background: Hepatitis B virus (HBV) infection is a major health concern with more than two billion individuals currently infected worldwide. Because of the limited effectiveness of existing vaccines and drugs, development of novel antiviral strategies is urgently needed. Heat stress cognate 70 (Hsc70) is an ATP-binding protein of the heat stress protein 70 family. Hsc70 has been found to be required for HBV DNA replication. Here we report, for the first time, that combined siRNAs targeting viral gene and siHsc70 are highly effective in suppressing ongoing HBV expression and replication.
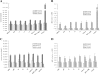
Methods: We constructed two plasmids (S1 and S2) expressing short hairpin RNAs (shRNAs) targeting surface open reading frame of HBV(HBVS) and one plasmid expressing shRNA targeting Hsc70 (siHsc70), and we used the EGFP-specific siRNA plasmid (siEGFP) as we had previously described. First, we evaluated the gene-silencing efficacy of both shRNAs using an enhanced green fluorescent protein (EGFP) reporter system and flow cytometry in HEK293 and T98G cells. Then, the antiviral potencies of HBV-specific siRNA (siHBV) in combination with siHsc70 in HepG2.2.15 cells were investigated. Moreover, type I IFN and TNF-α induction were measured by quantitative real-time PCR and ELISA.
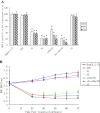
Results: Cotransfection of either S1 or S2 with an EGFP plasmid produced an 80%-90% reduction in EGFP signal relative to the control. This combinational RNAi effectively and specifically inhibited HBV protein, mRNA and HBV DNA, resulting in up to a 3.36 log10 reduction in HBV load in the HepG2.2.15 cell culture supernatants. The combined siRNAs were more potent than siHBV or siHsc70 used separately, and this approach can enhance potency in suppressing ongoing viral gene expression and replication in HepG2.2.15 cells while forestalling escape by mutant HBV. The antiviral synergy of siHBV used in combination with siHsc70 produced no cytotoxicity and induced no production of IFN-α, IFN-β and TNF-α in transfected cells.
Conclusions: Our combinational RNAi was sequence-specific, effective against wild-type and mutant drug-resistant HBV strains, without triggering interferon response or producing any side effects. These findings indicate that combinational RNAi has tremendous promise for developing innovative therapy against viral infection.
Figures


References
-
- Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. - PubMed
-
- Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395–403. - PubMed
-
- Ganem D, Prince AM. Hepatitis B virus infection-natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. - PubMed
-
- Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582–592. - PubMed
-
- Goldstein ST, Zhou F, Hadler SC, Bell BP, Mast EE, Margolis HS. A mathematical model to estimate global hepatitis B disease burden and vaccination impact. Int J Epidemiol. 2005;34:1329–1339. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

