Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome
- PMID: 23159369
- PMCID: PMC3508134
- DOI: 10.1016/j.cell.2012.09.045
Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome
Abstract
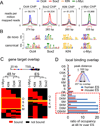
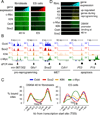
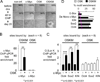
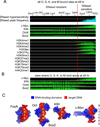
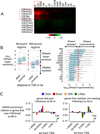
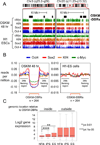
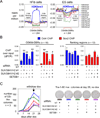
The ectopic expression of transcription factors can reprogram cell fate, yet it is unknown how the initial binding of factors to the genome relates functionally to the binding seen in the minority of cells that become reprogrammed. We report a map of Oct4, Sox2, Klf4, and c-Myc (O, S, K, and M) on the human genome during the first 48 hr of reprogramming fibroblasts to pluripotency. Three striking aspects of the initial chromatin binding events include an unexpected role for c-Myc in facilitating OSK chromatin engagement, the primacy of O, S, and K as pioneer factors at enhancers of genes that promote reprogramming, and megabase-scale chromatin domains spanned by H3K9me3, including many genes required for pluripotency, that prevent initial OSKM binding and impede the efficiency of reprogramming. We find diverse aspects of initial factor binding that must be overcome in the minority of cells that become reprogrammed.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
References
-
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

