Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma
- PMID: 23159910
- PMCID: PMC3717800
- DOI: 10.18632/oncotarget.709
Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma
Abstract
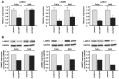
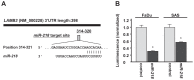
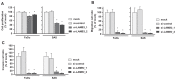
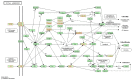
Recent our microRNA (miRNA) expression signature revealed that expression of microRNA-218 (miR-218) was reduced in cancer tissues, suggesting a candidate of tumor suppressor in head and neck squamous cell carcinoma (HNSCC). The aim of this study was to investigate the functional significance of miR-218 and its mediated moleculer pathways in HNSCC. Restoration of miR-218 in cancer cells led to significant inhibition of cell migration and invasion activities in HNSCC cell lines (FaDu and SAS). Genome-wide gene expression analysis of miR-218 transfectants and in silico database analysis showed that focal adhesion pathway was a promising candidate of miR-218 target pathways. The laminins are an important and biologically active part of the basal lamina, the function of that are various such as influencing cell differentiation, migration and adhesion as well as proliferation and cell survival. Interestingly, all components of laminin-332 (LAMA3, LAMB3 and LAMC2) are listed on the candidate genes in focal adhesion pathway. Furthermore, we focused on LAMB3 which has a miR-218 target site and gene expression studies and luciferase reporter assays showed that LAMB3 was directly regulated by miR-218. Silencing study of LAMB3 demonstrated significant inhibition of cell migration and invasion. In clinical specimens with HNSCC, the expression levels of laminin-332 were significantly upregulated in cancer tissues compared to adjacent non-cancerous tissues. Our analysis data showed that tumor suppressive miR-218 contributes to cancer cell migration and invasion through regulating focal adhesion pathway, especially laminin-332. Tumor suppressive miRNA-mediated novel cancer pathways provide new insights into the potential mechanisms of HNSCC oncogenesis.
Figures



References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. - PubMed
-
- Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Tran N, O'Brien CJ, Clark J, Rose B. Potential role of micro-RNAs in head and neck tumorigenesis. Head Neck. 2010;32:1099–1111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

