Peptidomic discovery of short open reading frame-encoded peptides in human cells
- PMID: 23160002
- PMCID: PMC3625679
- DOI: 10.1038/nchembio.1120
Peptidomic discovery of short open reading frame-encoded peptides in human cells
Abstract
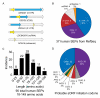
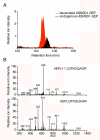
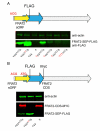
The complete extent to which the human genome is translated into polypeptides is of fundamental importance. We report a peptidomic strategy to detect short open reading frame (sORF)-encoded polypeptides (SEPs) in human cells. We identify 90 SEPs, 86 of which are previously uncharacterized, which is the largest number of human SEPs ever reported. SEP abundances range from 10-1,000 molecules per cell, identical to abundances of known proteins. SEPs arise from sORFs in noncoding RNAs as well as multicistronic mRNAs, and many SEPs initiate with non-AUG start codons, indicating that noncanonical translation may be more widespread in mammals than previously thought. In addition, coding sORFs are present in a small fraction (8 out of 1,866) of long intergenic noncoding RNAs. Together, these results provide strong evidence that the human proteome is more complex than previously appreciated.
Figures

Comment in
-
Genomics: Unknown polypeptides galore.Nat Methods. 2013 Jan;10(1):12. doi: 10.1038/nmeth.2321. Nat Methods. 2013. PMID: 23547289 No abstract available.
References
-
- Abastado JP, Miller PF, Hinnebusch AG. A quantitative model for translational control of the GCN4 gene of Saccharomyces cerevisiae. New Biol. 1991;3:511–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

