Profiling in situ microbial community structure with an amplification microarray
- PMID: 23160129
- PMCID: PMC3568579
- DOI: 10.1128/AEM.02664-12
Profiling in situ microbial community structure with an amplification microarray
Abstract
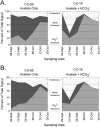
The objectives of this study were to unify amplification, labeling, and microarray hybridization chemistries within a single, closed microfluidic chamber (an amplification microarray) and verify technology performance on a series of groundwater samples from an in situ field experiment designed to compare U(VI) mobility under conditions of various alkalinities (as HCO(3)(-)) during stimulated microbial activity accompanying acetate amendment. Analytical limits of detection were between 2 and 200 cell equivalents of purified DNA. Amplification microarray signatures were well correlated with 16S rRNA-targeted quantitative PCR results and hybridization microarray signatures. The succession of the microbial community was evident with and consistent between the two microarray platforms. Amplification microarray analysis of acetate-treated groundwater showed elevated levels of iron-reducing bacteria (Flexibacter, Geobacter, Rhodoferax, and Shewanella) relative to the average background profile, as expected. Identical molecular signatures were evident in the transect treated with acetate plus NaHCO(3), but at much lower signal intensities and with a much more rapid decline (to nondetection). Azoarcus, Thaurea, and Methylobacterium were responsive in the acetate-only transect but not in the presence of bicarbonate. Observed differences in microbial community composition or response to bicarbonate amendment likely had an effect on measured rates of U reduction, with higher rates probable in the part of the field experiment that was amended with bicarbonate. The simplification in microarray-based work flow is a significant technological advance toward entirely closed-amplicon microarray-based tests and is generally extensible to any number of environmental monitoring applications.
Figures

Similar articles
-
Dynamic Succession of Groundwater Functional Microbial Communities in Response to Emulsified Vegetable Oil Amendment during Sustained In Situ U(VI) Reduction.Appl Environ Microbiol. 2015 Jun 15;81(12):4164-72. doi: 10.1128/AEM.00043-15. Epub 2015 Apr 10. Appl Environ Microbiol. 2015. PMID: 25862231 Free PMC article.
-
PCR amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip.Appl Environ Microbiol. 2011 Sep;77(18):6313-22. doi: 10.1128/AEM.05262-11. Epub 2011 Jul 15. Appl Environ Microbiol. 2011. PMID: 21764955 Free PMC article.
-
Monitoring microbial community structure and dynamics during in situ U(VI) bioremediation with a field-portable microarray analysis system.Environ Sci Technol. 2010 Jul 15;44(14):5516-22. doi: 10.1021/es1006498. Environ Sci Technol. 2010. PMID: 20560650
-
Permeable Reactive Barriers Designed To Mitigate Eutrophication Alter Bacterial Community Composition and Aquifer Redox Conditions.Appl Environ Microbiol. 2015 Oct;81(20):7114-24. doi: 10.1128/AEM.01986-15. Epub 2015 Jul 31. Appl Environ Microbiol. 2015. PMID: 26231655 Free PMC article.
-
Molecular analysis of the in situ growth rates of subsurface Geobacter species.Appl Environ Microbiol. 2013 Mar;79(5):1646-53. doi: 10.1128/AEM.03263-12. Epub 2012 Dec 28. Appl Environ Microbiol. 2013. PMID: 23275510 Free PMC article.
Cited by
-
Microbial Diversity in Deep-Subsurface Hot Brines of Northwest Poland: from Community Structure to Isolate Characteristics.Appl Environ Microbiol. 2020 May 5;86(10):e00252-20. doi: 10.1128/AEM.00252-20. Print 2020 May 5. Appl Environ Microbiol. 2020. PMID: 32198175 Free PMC article.
-
Application of Microfluidics in Experimental Ecology: The Importance of Being Spatial.Front Microbiol. 2018 Mar 20;9:496. doi: 10.3389/fmicb.2018.00496. eCollection 2018. Front Microbiol. 2018. PMID: 29616009 Free PMC article. Review.
-
The biogeochemical vertical structure renders a meromictic volcanic lake a trap for geogenic CO2 (Lake Averno, Italy).PLoS One. 2018 Mar 6;13(3):e0193914. doi: 10.1371/journal.pone.0193914. eCollection 2018. PLoS One. 2018. PMID: 29509779 Free PMC article.
References
-
- Handley KM, Wrighton KC, Piceno YM, Andersen GL, DeSantis TZ, Williams KH, Wilkins MJ, N′Guessan AL, Peacock A, Bargar J, Long PE, Banfield JF. 2012. High-density PhyloChip profiling of stimulated aquifer microbial communities reveals a complex response to acetate amendment. FEMS Microbiol. Ecol. 81:188–204 - PubMed
-
- Wilkins MJ, VerBerkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H, N′guessan AL, Thomas BC, Nicora CD, Shah MB, Abraham P, Lipton MS, Lovley DR, Hettich RL, Long PE, Banfield JF. 2009. Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 75:6591–6599 - PMC - PubMed
-
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665 - PubMed
-
- Waldron PJ, Wu L, Nostrand JDV, Schadt CW, He Z, Watson DB, Jardine PM, Palumbo AV, Hazen TC, Zhou J. 2009. Functional gene array-based analysis of microbial community structure in groundwaters with a gradient of contaminant levels. Environ. Sci. Technol. 43:3529–3534 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

