Evolutionary effects of translocations in bacterial genomes
- PMID: 23160175
- PMCID: PMC3542574
- DOI: 10.1093/gbe/evs099
Evolutionary effects of translocations in bacterial genomes
Abstract
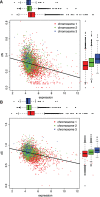
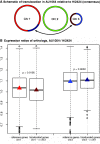
It has become clear that different genome regions need not evolve uniformly. This variation is particularly evident in bacterial genomes with multiple chromosomes, in which smaller, secondary chromosomes evolve more rapidly. We previously demonstrated that substitution rates and gene dispensability were greater on secondary chromosomes in many bacterial genomes. In Vibrio, the secondary chromosome is replicated later during the cell cycle, which reduces the effective dosage of these genes and hence their expression. More rapid evolution of secondary chromosomes may therefore reflect weaker purifying selection on less expressed genes. Here, we test this hypothesis by relating substitution rates of orthologs shared by multiple Burkholderia genomes, each with three chromosomes, to a study of gene expression in genomes differing by a major reciprocal translocation. This model predicts that expression should be greatest on chromosome 1 (the largest) and least on chromosome 3 (the smallest) and that expression should tend to decline within chromosomes from replication origin to terminus. Moreover, gene movement to the primary chromosome should associate with increased expression, and movement to secondary chromosomes should result in reduced expression. Our analysis supports each of these predictions, as translocated genes tended to shift expression toward their new chromosome neighbors despite inevitable cis-acting regulation of expression. This study sheds light on the early dynamics of genomes following rearrangement and illustrates how secondary chromosomes in bacteria may become evolutionary test beds.
Figures
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Couturier E, Rocha EP. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol. 2006;59:1506–1518. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

