Evolutionary rate and duplicability in the Arabidopsis thaliana protein-protein interaction network
- PMID: 23160177
- PMCID: PMC3542556
- DOI: 10.1093/gbe/evs101
Evolutionary rate and duplicability in the Arabidopsis thaliana protein-protein interaction network
Abstract
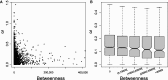
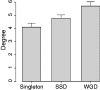
Genes show a bewildering variation in their patterns of molecular evolution, as a result of the action of different levels and types of selective forces. The factors underlying this variation are, however, still poorly understood. In the last decade, the position of proteins in the protein-protein interaction network has been put forward as a determinant factor of the evolutionary rate and duplicability of their encoding genes. This conclusion, however, has been based on the analysis of the limited number of microbes and animals for which interactome-level data are available (essentially, Escherichia coli, yeast, worm, fly, and humans). Here, we study, for the first time, the relationship between the position of proteins in the high-density interactome of a plant (Arabidopsis thaliana) and the patterns of molecular evolution of their encoding genes. We found that genes whose encoded products act at the center of the network are more evolutionarily constrained than those acting at the network periphery. This trend remains significant when potential confounding factors (gene expression level and breadth, duplicability, function, and length of the encoded products) are controlled for. Even though the correlation between centrality measures and rates of evolution is generally weak, for some functional categories, it is comparable in strength to (or even stronger than) the correlation between evolutionary rates and expression levels or breadths. In addition, genes encoding interacting proteins in the network evolve at relatively similar rates. Finally, Arabidopsis proteins encoded by duplicated genes are more highly connected than those encoded by singleton genes. This observation is in agreement with the patterns observed in humans, but in contrast with those observed in E. coli, yeast, worm, and fly (whose duplicated genes tend to act at the periphery of the network), implying that the relationship between duplicability and centrality inverted at least twice during eukaryote evolution. Taken together, these results indicate that the structure of the A. thaliana network constrains the evolution of its components at multiple levels.
Figures
References
-
- Albert R, Jeong H, Barabási AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

