Identification of a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with multiple sclerosis
- PMID: 23160276
- PMCID: PMC3538279
- DOI: 10.1136/jmedgenet-2012-101085
Identification of a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with multiple sclerosis
Abstract
Background and aim: Several studies have highlighted the association of the 12q13.3-12q14.1 region with coeliac disease, type 1 diabetes, rheumatoid arthritis and multiple sclerosis (MS); however, the causal variants underlying diseases are still unclear. The authors sought to identify the functional variant of this region associated with MS.
Methods: Tag-single nucleotide polymorphism (SNP) analysis of the associated region encoding 15 genes was performed in 2876 MS patients and 2910 healthy Caucasian controls together with expression regulation analyses.
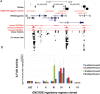
Results: rs6581155, which tagged 18 variants within a region where 9 genes map, was sufficient to model the association. This SNP was in total linkage disequilibrium (LD) with other polymorphisms that associated with the expression levels of FAM119B, AVIL, TSFM, TSPAN31 and CYP27B1 genes in different expression quantitative trait loci studies. Functional annotations from Encyclopedia of DNA Elements (ENCODE) showed that six out of these rs6581155-tagged-SNPs were located in regions with regulatory potential and only one of them, rs10877013, exhibited allele-dependent (ratio A/G=9.5-fold) and orientation-dependent (forward/reverse=2.7-fold) enhancer activity as determined by luciferase reporter assays. This enhancer is located in a region where a long-range chromatin interaction among the promoters and promoter-enhancer of several genes has been described, possibly affecting their expression simultaneously.
Conclusions: This study determines a functional variant which alters the enhancer activity of a regulatory element in the locus affecting the expression of several genes and explains the association of the 12q13.3-12q14.1 region with MS.
Figures


References
-
- Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 2009;52:61–76 - PubMed
-
- McElroy JP, Oksenberg JR. Multiple sclerosis genetics. Curr Top Microbiol Immunol 2008;318:45–72 - PubMed
-
- Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Curr Opin Neurol 2009;22:219–25 - PubMed
-
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 2009;461:747–53 - PMC - PubMed
