MxA transcripts with distinct first exons and modulation of gene expression levels by single-nucleotide polymorphisms in human bronchial epithelial cells
- PMID: 23160781
- PMCID: PMC7079882
- DOI: 10.1007/s00251-012-0663-8
MxA transcripts with distinct first exons and modulation of gene expression levels by single-nucleotide polymorphisms in human bronchial epithelial cells
Abstract
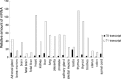
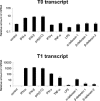
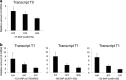
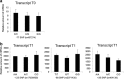
Myxovirus resistance A (MxA) is a major interferon (IFN)-inducible antiviral protein. Promoter single-nucleotide polymorphisms (SNPs) of MxA near the IFN-stimulated response element (ISRE) have been frequently associated with various viral diseases, including emerging respiratory infections. We investigated the expression profile of MxA transcripts with distinct first exons in human bronchial epithelial cells. For primary culture, the bronchial epithelium was isolated from lung tissues with different genotypes, and total RNA was subjected to real-time reverse transcription polymerase chain reaction. The previously reported MxA transcript (T1) and a recently registered transcript with a distinct 5' first exon (T0) were identified. IFN-β and polyinosinic-polycytidylic acid induced approximately 100-fold higher expression of the T1 transcript than that of the T0 transcript, which also had a potential ISRE motif near its transcription start site. Even without inducers, the T1 transcript accounted for approximately two thirds of the total expression of MxA, levels of which were significantly associated with its promoter and exon 1 SNPs (rs17000900, rs2071430, and rs464138). Our results suggest that MxA observed in respiratory viral infections is possibly dominated by the T1 transcript and partly influenced by relevant 5' SNPs.
Figures

Similar articles
-
The myxovirus resistance A (MxA) gene -88G>T single nucleotide polymorphism is associated with prostate cancer.Infect Genet Evol. 2013 Jun;16:186-90. doi: 10.1016/j.meegid.2013.02.010. Epub 2013 Feb 22. Infect Genet Evol. 2013. PMID: 23438650 Free PMC article.
-
Pharmacogenetics of MXA SNPs in interferon-beta treated multiple sclerosis patients.J Neuroimmunol. 2007 Jan;182(1-2):236-9. doi: 10.1016/j.jneuroim.2006.10.011. Epub 2006 Nov 27. J Neuroimmunol. 2007. PMID: 17126411
-
Differential transcriptional expresión of the polymorphic myxovirus resistance protein A in response to interferon-alpha treatment.Pharmacogenetics. 2004 Mar;14(3):189-93. doi: 10.1097/00008571-200403000-00007. Pharmacogenetics. 2004. PMID: 15167707 Clinical Trial.
-
Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity.J Interferon Cytokine Res. 2011 Jan;31(1):79-87. doi: 10.1089/jir.2010.0076. Epub 2010 Dec 19. J Interferon Cytokine Res. 2011. PMID: 21166595 Review.
-
[Alpha interferon, antiviral proteins and their value in clinical medicine].Ann Biol Clin (Paris). 1999 Nov-Dec;57(6):659-66. Ann Biol Clin (Paris). 1999. PMID: 10572214 Review. French.
Cited by
-
The myxovirus resistance A (MxA) gene -88G>T single nucleotide polymorphism is associated with prostate cancer.Infect Genet Evol. 2013 Jun;16:186-90. doi: 10.1016/j.meegid.2013.02.010. Epub 2013 Feb 22. Infect Genet Evol. 2013. PMID: 23438650 Free PMC article.
-
Myxovirus resistance protein A (MxA) polymorphism is associated with IFNβ response in Iranian multiple sclerosis patients.Neurol Sci. 2017 Jun;38(6):1093-1099. doi: 10.1007/s10072-017-2935-4. Epub 2017 Apr 6. Neurol Sci. 2017. PMID: 28386647
References
-
- Abe H, Hayes CN, Ochi H, Maekawa T, Tsuge M, Miki D, Mitsui F, Hiraga N, Imamura M, Takahashi S, Kubo M, Nakamura Y, Chayama K. IL28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. J Hepatol. 2011;54:1094–1101. doi: 10.1016/j.jhep.2010.09.019. - DOI - PubMed
-
- Ching JC, Chan KY, Lee EH, Xu MS, Ting CK, So TM, Sham PC, Leung GM, Peiris JS, Khoo US. Significance of the myxovirus resistance A (MxA) gene −123C > a single-nucleotide polymorphism in suppressed interferon beta induction of severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2010;201:1899–1908. doi: 10.1086/652799. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

