miRNA repression of translation in vitro takes place during 43S ribosomal scanning
- PMID: 23161679
- PMCID: PMC3592420
- DOI: 10.1093/nar/gks1076
miRNA repression of translation in vitro takes place during 43S ribosomal scanning
Abstract
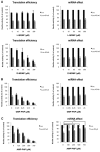
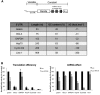
microRNAs (miRNAs) regulate gene expression at multiple levels by repressing translation, stimulating deadenylation and inducing the premature decay of target messenger RNAs (mRNAs). Although the mechanism by which miRNAs repress translation has been widely studied, the precise step targeted and the molecular insights of such repression are still evasive. Here, we have used our newly designed in vitro system, which allows to study miRNA effect on translation independently of deadenylation. By using specific inhibitors of various stages of protein synthesis, we first show that miRNAs target exclusively the early steps of translation with no effect on 60S ribosomal subunit joining, elongation or termination. Then, by using viral proteases and IRES-driven mRNA constructs, we found that translational inhibition takes place during 43S ribosomal scanning and requires both the poly(A) binding protein and eIF4G independently from their physical interaction.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

