Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation
- PMID: 23161683
- PMCID: PMC3592403
- DOI: 10.1093/nar/gks1071
Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation
Abstract
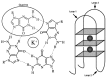
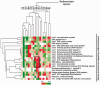
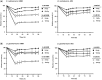
A remarkable number of guanine-rich sequences with potential to adopt non-canonical secondary structures called G-quadruplexes (or G4 DNA) are found within gene promoters. Despite growing interest, regulatory role of quadruplex DNA motifs in intrinsic cellular function remains poorly understood. Herein, we asked whether occurrence of potential G4 (PG4) DNA in promoters is associated with specific function(s) in bacteria. Using a normalized promoter-PG4-content (PG4(P)) index we analysed >60,000 promoters in 19 well-annotated species for (a) function class(es) and (b) gene(s) with enriched PG4(P). Unexpectedly, PG4-associated functional classes were organism specific, suggesting that PG4 motifs may impart specific function to organisms. As a case study, we analysed radioresistance. Interestingly, unsupervised clustering using PG4(P) of 21 genes, crucial for radioresistance, grouped three radioresistant microorganisms including Deinococcus radiodurans. Based on these predictions we tested and found that in presence of nanomolar amounts of the intracellular quadruplex-binding ligand N-methyl mesoporphyrin (NMM), radioresistance of D. radiodurans was attenuated by ~60%. In addition, important components of the RecF recombinational repair pathway recA, recF, recO, recR and recQ genes were found to harbour promoter-PG4 motifs and were also down-regulated in presence of NMM. Together these results provide first evidence that radioresistance may involve G4 DNA-mediated regulation and support the rationale that promoter-PG4s influence selective functions.
Figures




References
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. - PubMed
-
- Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

