Contribution of intersubunit bridges to the energy barrier of ribosomal translocation
- PMID: 23161696
- PMCID: PMC3592451
- DOI: 10.1093/nar/gks1074
Contribution of intersubunit bridges to the energy barrier of ribosomal translocation
Abstract
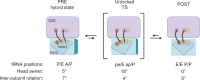
In every round of translation elongation, EF-G catalyzes translocation, the movement of tRNAs (and paired codons) to their adjacent binding sites in the ribosome. Previous kinetic studies have shown that the rate of tRNA-mRNA movement is limited by a conformational change in the ribosome termed 'unlocking'. Although structural studies offer some clues as to what unlocking might entail, the molecular basis of this conformational change remains an open question. In this study, the contribution of intersubunit bridges to the energy barrier of translocation was systematically investigated. Unlike those targeting B2a and B3, mutations that disrupt bridges B1a, B4, B7a and B8 increased the maximal rate of both forward (EF-G dependent) and reverse (spontaneous) translocation. As bridge B1a is predicted to constrain 30S head movement and B4, B7a and B8 are predicted to constrain intersubunit rotation, these data provide evidence that formation of the unlocked (transition) state involves both 30S head movement and intersubunit rotation.
Figures


References
-
- Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Let. 1971;17:324–326. - PubMed
-
- Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969;244:1533–1539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

