A broad specificity nucleoside kinase from Thermoplasma acidophilum
- PMID: 23161756
- PMCID: PMC3595323
- DOI: 10.1002/prot.24212
A broad specificity nucleoside kinase from Thermoplasma acidophilum
Abstract
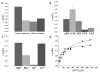
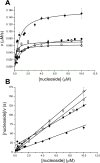
The crystal structure of Ta0880, determined at 1.91 Å resolution, from Thermoplasma acidophilum revealed a dimer with each monomer composed of an α/β/α sandwich domain and a smaller lid domain. The overall fold belongs to the PfkB family of carbohydrate kinases (a family member of the Ribokinase clan) which include ribokinases, 1-phosphofructokinases, 6-phosphofructo-2-kinase, inosine/guanosine kinases, fructokinases, adenosine kinases, and many more. Based on its general fold, Ta0880 had been annotated as a ribokinase-like protein. Using a coupled pyruvate kinase/lactate dehydrogenase assay, the activity of Ta0880 was assessed against a variety of ribokinase/pfkB-like family substrates; activity was not observed for ribose, fructose-1-phosphate, or fructose-6-phosphate. Based on structural similarity with nucleoside kinases (NK) from Methanocaldococcus jannaschii (MjNK, PDB 2C49, and 2C4E) and Burkholderia thailandensis (BtNK, PDB 3B1O), nucleoside kinase activity was investigated. Ta0880 (TaNK) was confirmed to have nucleoside kinase activity with an apparent KM for guanosine of 0.21 μM and catalytic efficiency of 345,000 M(-1) s(-1) . These three NKs have significantly different substrate, phosphate donor, and cation specificities and comparisons of specificity and structure identified residues likely responsible for the nucleoside substrate selectivity. Phylogenetic analysis identified three clusters within the PfkB family and indicates that TaNK is a member of a new sub-family with broad nucleoside specificities. Proteins 2013. © 2012 Wiley Periodicals, Inc.
Copyright © 2012 Wiley Periodicals, Inc.
Figures







Similar articles
-
Structure of Methanocaldococcus jannaschii nucleoside kinase: an archaeal member of the ribokinase family.Acta Crystallogr D Biol Crystallogr. 2006 Sep;62(Pt 9):1085-97. doi: 10.1107/S0907444906024826. Epub 2006 Aug 19. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16929110
-
A novel nucleoside kinase from Burkholderia thailandensis: a member of the phosphofructokinase B-type family of enzymes.FEBS J. 2008 Dec;275(23):5865-72. doi: 10.1111/j.1742-4658.2008.06716.x. FEBS J. 2008. PMID: 19021762
-
The phosphofructokinase-B (MJ0406) from Methanocaldococcus jannaschii represents a nucleoside kinase with a broad substrate specificity.Extremophiles. 2007 Jan;11(1):105-14. doi: 10.1007/s00792-006-0018-1. Epub 2006 Oct 5. Extremophiles. 2007. PMID: 17021658
-
Expression, purification, crystallization and preliminary X-ray analysis of a nucleoside kinase from the hyperthermophile Methanocaldococcus jannaschii.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005 Jun 1;61(Pt 6):591-4. doi: 10.1107/S1744309105015642. Epub 2005 Jun 1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005. PMID: 16511104 Free PMC article.
-
Catalytic mechanism of the 20S proteasome of Thermoplasma acidophilum revealed by X-ray crystallography.Cold Spring Harb Symp Quant Biol. 1995;60:525-32. doi: 10.1101/sqb.1995.060.01.056. Cold Spring Harb Symp Quant Biol. 1995. PMID: 8824425 Review. No abstract available.
Cited by
-
Objective: biochemical function.Front Genet. 2014 Jul 8;5:210. doi: 10.3389/fgene.2014.00210. eCollection 2014. Front Genet. 2014. PMID: 25071837 Free PMC article. No abstract available.
-
A Phosphofructokinase Homolog from Pyrobaculum calidifontis Displays Kinase Activity towards Pyrimidine Nucleosides and Ribose 1-Phosphate.J Bacteriol. 2018 Jul 25;200(16):e00284-18. doi: 10.1128/JB.00284-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29866806 Free PMC article.
-
A pentose bisphosphate pathway for nucleoside degradation in Archaea.Nat Chem Biol. 2015 May;11(5):355-60. doi: 10.1038/nchembio.1786. Epub 2015 Mar 30. Nat Chem Biol. 2015. PMID: 25822915
-
Multifactorial interaction of selenium, iron, xylose, and glycine on cordycepin metabolism in Cordyceps militaris.Appl Microbiol Biotechnol. 2023 Dec;107(24):7403-7416. doi: 10.1007/s00253-023-12792-x. Epub 2023 Sep 29. Appl Microbiol Biotechnol. 2023. PMID: 37773218
-
The COMBREX project: design, methodology, and initial results.PLoS Biol. 2013;11(8):e1001638. doi: 10.1371/journal.pbio.1001638. Epub 2013 Aug 27. PLoS Biol. 2013. PMID: 24013487 Free PMC article.
References
-
- Nyhan WL. Encyclopedia of Life Sciences. John Wiley & Sons; 2005. Nucloside synthesis via salvage pathways; pp. 1–7.
-
- Hansen T, Arnfors L, Ladenstein R, Schonheit P. The phosphofructokinase-B (MJ0406) from Methanocaldococcus jannaschii represents a nucleoside kinase with a broad substrate specificity. Extremophiles. 2007;11:105–114. - PubMed
-
- Ota H, Sakasegawa S, Yasuda Y, Imamura S, Tamura T. A novel nucleoside kinase from Burkholderia thailandensis: a member of the phosphofructokinase B-type family of enzymes. FEBS J. 2008;275:5865–5872. - PubMed
-
- Hansen T, Schonheit P. Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a non-allosteric enzyme related to the phosphofructokinase-B sugar kinase family, from the hyperthermophilic crenarchaeote Aeropyrum pernix. Arch Microbiol. 2001;177:62–69. - PubMed
-
- Arnfors L, Hansen T, Schonheit P, Ladenstein R, Meining W. Structure of Methanocaldococcus jannaschii nucleoside kinase: an archaeal member of the ribokinase family. Acta Crystallogr D Biol Crystallogr. 2006;62:1085–1097. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

