Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies
- PMID: 23161787
- PMCID: PMC3635509
- DOI: 10.1093/neuonc/nos284
Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies
Abstract
Background: Genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) at 7 loci influencing glioma risk: rs2736100 (TERT), rs11979158 and rs2252586 (EGFR), rs4295627 (CCDC26), rs4977756 (CDKN2A/CDKN2B), rs498872 (PHLDB1), and rs6010620 (RTEL1).
Materials and methods: We studied the relationship among these 7 glioma-risk SNPs and characteristics of tumors from 1374 patients, including grade, IDH (ie IDH1 or IDH2) mutation, EGFR amplification, CDKN2A-p16-INK4a homozygous deletion, 9p and 10q loss, and 1p-19q codeletion.
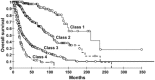
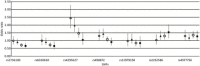
Results: rs2736100 (TERT) and rs6010620 (RTEL1) risk alleles were associated with high-grade disease, EGFR amplification, CDKN2A-p16-INK4a homozygous deletion, and 9p and 10q deletion; rs4295627 (CCDC26) and rs498872 (PHLDB1) were associated with low-grade disease, IDH mutation, and 1p-19q codeletion. In contrast, rs4977756 (CDKN2A/B), rs11979158 (EGFR), and to a lesser extent, rs2252586 (EGFR) risk alleles were independent of tumor grade and genetic profile. Adjusting for tumor grade showed a significant association between rs2736100 and IDH status (P = .01), 10q loss (P = .02); rs4295627 and 1p-19q codeletion (P = .04), rs498872 and IDH (P = .02), 9p loss (P = .04), and 10q loss (P = .02). Case-control analyses stratified into 4 molecular classes (defined by 1p-19q status, IDH mutation, and EGFR amplification) showed an association of rs4295627 and rs498872 with IDH-mutated gliomas (P < 10(-3)) and rs2736100 and rs6010620 with IDH wild-type gliomas (P < 10(-3) and P = .03).
Conclusion: The frequency of EGFR and CDKN2A/B risk alleles were largely independent of tumor genetic profile, whereas TERT, RTEL1, CCDC26, and PHLDB1 variants were associated with different genetic profiles that annotate distinct molecular pathways. Our findings provide further insight into the biological basis of glioma etiology.
Figures
Comment in
-
From GWAS risk foci to glioma molecular subclass.Neuro Oncol. 2013 May;15(5):513-4. doi: 10.1093/neuonc/not061. Neuro Oncol. 2013. PMID: 23620433 Free PMC article. No abstract available.
Similar articles
-
Genetic risk variants in the CDKN2A/B, RTEL1 and EGFR genes are associated with somatic biomarkers in glioma.J Neurooncol. 2016 May;127(3):483-92. doi: 10.1007/s11060-016-2066-4. Epub 2016 Feb 2. J Neurooncol. 2016. PMID: 26839018 Free PMC article.
-
Replication of GWAS identifies RTEL1, CDKN2A/B, and PHLDB1 SNPs as risk factors in Portuguese gliomas patients.Mol Biol Rep. 2020 Feb;47(2):877-886. doi: 10.1007/s11033-019-05178-8. Epub 2019 Nov 12. Mol Biol Rep. 2020. PMID: 31721021
-
Genetic risk profiles identify different molecular etiologies for glioma.Clin Cancer Res. 2010 Nov 1;16(21):5252-9. doi: 10.1158/1078-0432.CCR-10-1502. Epub 2010 Sep 16. Clin Cancer Res. 2010. PMID: 20847058 Free PMC article.
-
Genetic causes of glioma: new leads in the labyrinth.Curr Opin Oncol. 2011 Nov;23(6):643-7. doi: 10.1097/CCO.0b013e32834a6f61. Curr Opin Oncol. 2011. PMID: 21825990 Review.
-
Pertinence of glioma and single nucleotide polymorphism of TERT, CCDC26, CDKN2A/B and RTEL1 genes in glioma: a meta-analysis.Front Oncol. 2023 Sep 7;13:1180099. doi: 10.3389/fonc.2023.1180099. eCollection 2023. Front Oncol. 2023. PMID: 37746290 Free PMC article.
Cited by
-
Telomere Maintenance Mechanisms in Cancer.Genes (Basel). 2018 May 3;9(5):241. doi: 10.3390/genes9050241. Genes (Basel). 2018. PMID: 29751586 Free PMC article. Review.
-
IDH wild-type lower-grade gliomas with glioblastoma molecular features: a systematic review and meta-analysis.Brain Tumor Pathol. 2023 Jul;40(3):143-157. doi: 10.1007/s10014-023-00463-8. Epub 2023 May 22. Brain Tumor Pathol. 2023. PMID: 37212969
-
Methylation of the miR-126 gene associated with glioma progression.Fam Cancer. 2016 Apr;15(2):317-24. doi: 10.1007/s10689-015-9846-4. Fam Cancer. 2016. PMID: 26463235
-
TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations.Br J Cancer. 2014 Nov 11;111(10):2024-32. doi: 10.1038/bjc.2014.538. Epub 2014 Oct 14. Br J Cancer. 2014. PMID: 25314060 Free PMC article.
-
Genetics in glioma: lessons learned from genome-wide association studies.Curr Opin Neurol. 2013 Dec;26(6):688-92. doi: 10.1097/WCO.0000000000000033. Curr Opin Neurol. 2013. PMID: 24184969 Free PMC article. Review.
References
-
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics. CA Cancer J Clin. 2011;61:212–236. - PubMed
-
- Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. - PubMed
-
- Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74:1886–1890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

