Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6
- PMID: 23161908
- PMCID: PMC3528572
- DOI: 10.1073/pnas.1203035109
Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6
Abstract
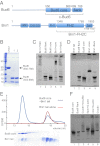
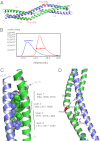
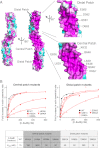
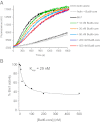
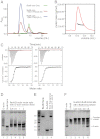
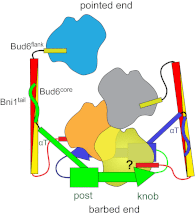
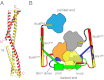
Formin proteins and their associated factors cooperate to assemble unbranched actin filaments in diverse cellular structures. The Saccharomyces cerevisiae formin Bni1 and its associated nucleation-promoting factor (NPF) Bud6 generate actin cables and mediate polarized cell growth. Bud6 binds to both the tail of the formin and G-actin, thereby recruiting monomeric actin to the formin to create a nucleation seed. Here, we structurally and functionally dissect the nucleation-promoting C-terminal region of Bud6 into a Bni1-binding "core" domain and a G-actin binding "flank" domain. The ∼2-Å resolution crystal structure of the Bud6 core domain reveals an elongated dimeric rod with a unique fold resembling a triple-helical coiled-coil. Binding and actin-assembly assays show that conserved residues on the surface of this domain mediate binding to Bni1 and are required for NPF activity. We find that the Bni1 dimer binds two Bud6 dimers and that the Bud6 flank binds a single G-actin molecule. These findings suggest a model in which a Bni1/Bud6 complex with a 2:4 subunit stoichiometry assembles a nucleation seed with Bud6 coordinating up to four actin subunits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif.Structure. 2015 Aug 4;23(8):1492-1499. doi: 10.1016/j.str.2015.05.015. Epub 2015 Jun 25. Structure. 2015. PMID: 26118535 Free PMC article.
-
Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor.Mol Biol Cell. 2011 Nov;22(21):4016-28. doi: 10.1091/mbc.E11-05-0404. Epub 2011 Aug 31. Mol Biol Cell. 2011. PMID: 21880892 Free PMC article.
-
Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6.J Biol Chem. 2005 Jul 29;280(30):28023-33. doi: 10.1074/jbc.M503094200. Epub 2005 May 27. J Biol Chem. 2005. PMID: 15923184
-
Formins: signaling effectors for assembly and polarization of actin filaments.J Cell Sci. 2003 Jul 1;116(Pt 13):2603-11. doi: 10.1242/jcs.00611. J Cell Sci. 2003. PMID: 12775772 Review.
-
The ADF homology (ADF-H) domain: a highly exploited actin-binding module.Mol Biol Cell. 1998 Aug;9(8):1951-9. doi: 10.1091/mbc.9.8.1951. Mol Biol Cell. 1998. PMID: 9693358 Free PMC article. Review. No abstract available.
Cited by
-
A time-resolved interaction analysis of Bem1 reconstructs the flow of Cdc42 during polar growth.Life Sci Alliance. 2020 Jul 31;3(9):e202000813. doi: 10.26508/lsa.202000813. Print 2020 Sep. Life Sci Alliance. 2020. PMID: 32737079 Free PMC article.
-
Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism.Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):E2677-86. doi: 10.1073/pnas.1307235110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818602 Free PMC article.
-
A teamwork promotion of formin-mediated actin nucleation by Bud6 and Aip5 in Saccharomyces cerevisiae.Mol Biol Cell. 2022 Feb 1;33(2):ar19. doi: 10.1091/mbc.E21-06-0285. Epub 2021 Nov 24. Mol Biol Cell. 2022. PMID: 34818061 Free PMC article.
-
Orchestrated actin nucleation by the Candida albicans polarisome complex enables filamentous growth.J Biol Chem. 2020 Oct 30;295(44):14840-14854. doi: 10.1074/jbc.RA120.013890. Epub 2020 Aug 26. J Biol Chem. 2020. PMID: 32848016 Free PMC article.
-
Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation.J Cell Biol. 2013 May 13;201(4):595-611. doi: 10.1083/jcb.201212059. J Cell Biol. 2013. PMID: 23671312 Free PMC article.
References
-
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

