Cancer metabolism: what we can learn from proteomic analysis by mass spectrometry
- PMID: 23162076
- PMCID: PMC5547437
Cancer metabolism: what we can learn from proteomic analysis by mass spectrometry
Abstract
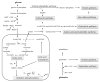
A variety of genomic and proteomic tools have been used to study cancer metabolism and metabolomics in order to understand how cancer cells survive in their environment. Throughout the past decade, mass spectrometry has been routinely used for large-scale protein identification of complex biological mixtures. In this review, we discuss some recent developments in cancer metabolism by proteomic analysis using mass spectrometric techniques, focusing on pyruvate kinase, L-lactate dehydrogenase, Warburg effect, glutamine metabolism and oxidative stress.
Figures
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Garland Science. Taylor & Francis Group; New York: 2007. Molecular Biology of the Cell.
-
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. - PubMed
-
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. - PubMed
-
- Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
