Sequencing of chloroplast genome using whole cellular DNA and solexa sequencing technology
- PMID: 23162558
- PMCID: PMC3492724
- DOI: 10.3389/fpls.2012.00243
Sequencing of chloroplast genome using whole cellular DNA and solexa sequencing technology
Abstract
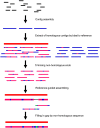
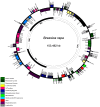
Sequencing of the chloroplast (cp) genome using traditional sequencing methods has been difficult because of its size (>120 kb) and the complicated procedures required to prepare templates. To explore the feasibility of sequencing the cp genome using DNA extracted from whole cells and Solexa sequencing technology, we sequenced whole cellular DNA isolated from leaves of three Brassicarapa accessions with one lane per accession. In total, 246, 362, and 361 Mb sequence data were generated for the three accessions Chiifu-401-42, Z16, and FT, respectively. Micro-reads were assembled by reference-guided assembly using the cpDNA sequences of B. rapa, Arabidopsis thaliana, and Nicotiana tabacum. We achieved coverage of more than 99.96% of the cp genome in the three tested accessions using the B. rapa sequence as the reference. When A. thaliana or N. tabacum sequences were used as references, 99.7-99.8 or 95.5-99.7% of the B. rapa cp genome was covered, respectively. These results demonstrated that sequencing of whole cellular DNA isolated from young leaves using the Illumina Genome Analyzer is an efficient method for high-throughput sequencing of cp genome.
Keywords: Brassica rapa; Solexa sequencing technology; chloroplast genome; sequencing; whole cellular DNA.
Figures


References
-
- Fulton T. M., Chunwongse J., Tanksley S. D. (1995). Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13, 207–20910.1007/BF02670897 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

