The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP
- PMID: 23163901
- PMCID: PMC5034864
- DOI: 10.1111/mmi.12066
The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP
Abstract
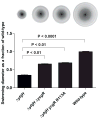
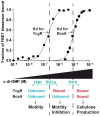
c-di-GMP is a bacterial second messenger that is enzymatically synthesized and degraded in response to environmental signals. Cellular processes are affected when c-di-GMP binds to receptors which include proteins that contain the PilZ domain. Although each c-di-GMP synthesis or degradation enzyme metabolizes the same molecule, many of these enzymes can be linked to specific downstream processes. Here we present evidence that c-di-GMP signalling specificity is achieved through differences in affinities of receptor macromolecules. We show that the PilZ domain proteins of Salmonella Typhimurium, YcgR and BcsA, demonstrate a 43-fold difference in their affinity for c-di-GMP. Modulation of the affinities of these proteins altered their activities in a predictable manner in vivo. Inactivation of yhjH, which encodes a predicted c-di-GMP degrading enzyme, increased the fraction of the cellular population that demonstrated c-di-GMP levels high enough to bind to the higher-affinity YcgR protein and inhibit motility, but not high enough to bind to the lower-affinity BcsA protein and stimulate cellulose production. Finally, PilZ domain proteins of Pseudomonas aeruginosa demonstrated a 145-fold difference in binding affinities, suggesting that regulation by binding affinity may be a conserved mechanism that allows organisms with many c-di-GMP binding macromolecules to rapidly integrate multiple environmental signals into one output.
© 2012 Blackwell Publishing Ltd.
Figures






References
-
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. - PubMed
-
- Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–116. - PubMed
-
- Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–32024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

