Regulated protein aggregation: stress granules and neurodegeneration
- PMID: 23164372
- PMCID: PMC3519755
- DOI: 10.1186/1750-1326-7-56
Regulated protein aggregation: stress granules and neurodegeneration
Abstract
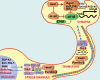
The protein aggregation that occurs in neurodegenerative diseases is classically thought to occur as an undesirable, nonfunctional byproduct of protein misfolding. This model contrasts with the biology of RNA binding proteins, many of which are linked to neurodegenerative diseases. RNA binding proteins use protein aggregation as part of a normal regulated, physiological mechanism controlling protein synthesis. The process of regulated protein aggregation is most evident in formation of stress granules. Stress granules assemble when RNA binding proteins aggregate through their glycine rich domains. Stress granules function to sequester, silence and/or degrade RNA transcripts as part of a mechanism that adapts patterns of local RNA translation to facilitate the stress response. Aggregation of RNA binding proteins is reversible and is tightly regulated through pathways, such as phosphorylation of elongation initiation factor 2α. Microtubule associated protein tau also appears to regulate stress granule formation. Conversely, stress granule formation stimulates pathological changes associated with tau. In this review, I propose that the aggregation of many pathological, intracellular proteins, including TDP-43, FUS or tau, proceeds through the stress granule pathway. Mutations in genes coding for stress granule associated proteins or prolonged physiological stress, lead to enhanced stress granule formation, which accelerates the pathophysiology of protein aggregation in neurodegenerative diseases. Over-active stress granule formation could act to sequester functional RNA binding proteins and/or interfere with mRNA transport and translation, each of which might potentiate neurodegeneration. The reversibility of the stress granule pathway also offers novel opportunities to stimulate endogenous biochemical pathways to disaggregate these pathological stress granules, and perhaps delay the progression of disease.
Figures
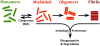


References
-
- Jarrett J, Jarrett J, Lansbury P. Seeding "one-dimensional crystallization" of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73(P Lansbury):1055–1058. - PubMed
-
- Bandhyopadhyay U, Cuervo AM. Chaperone-mediated autophagy in aging and neurodegeneration: lessons from alpha-synuclein. Exp Gerontol. 2006;42:120–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

