Proteomic and functional analyses of protein-DNA complexes during gene transfer
- PMID: 23164933
- PMCID: PMC3616537
- DOI: 10.1038/mt.2012.231
Proteomic and functional analyses of protein-DNA complexes during gene transfer
Abstract
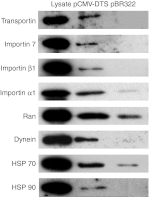
One of the barriers to successful nonviral gene delivery is the crowded cytoplasm, which plasmids need to actively traverse for gene expression. Relatively little is known about how this process occurs, but our lab and others have shown that the microtubule network and motors are required for plasmid movement to the nucleus. To further investigate how plasmids exploit normal physiological processes to transfect cells, we have taken a proteomics approach to identify the proteins that comprise the plasmid-trafficking complex. We have developed a live cell DNA-protein pull-down assay to isolate complexes at certain time points post-transfection (15 minutes to 4 hours) for analysis by mass spectrometry (MS). Plasmids containing promoter sequences bound hundreds of unique proteins as early as 15 minutes post-electroporation, while a plasmid lacking any eukaryotic sequences failed to bind many of the proteins. Specific proteins included microtubule-based motor proteins (e.g., kinesin and dynein), proteins involved in protein nuclear import (e.g., importin 1, 2, 4, and 7, Crm1, RAN, and several RAN-binding proteins), a number of heterogeneous nuclear ribonucleoprotein (hnRNP)- and mRNA-binding proteins, and transcription factors. The significance of several of the proteins involved in protein nuclear localization and plasmid trafficking was determined by monitoring movement of microinjected fluorescently labeled plasmids via live cell particle tracking in cells following protein knockdown by small-interfering RNA (siRNA) or through the use of specific inhibitors. While importin β1 was required for plasmid trafficking and subsequent nuclear import, importin α1 played no role in microtubule trafficking but was required for optimal plasmid nuclear import. Surprisingly, the nuclear export protein Crm1 also was found to complex with the transfected plasmids and was necessary for plasmid trafficking along microtubules and nuclear import. Our results show that various proteins involved in nuclear import and export influence intracellular trafficking of plasmids and subsequent nuclear accumulation.
Figures
References
-
- Mesika A, Kiss V, Brumfeld V, Ghosh G., and, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

