Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53
- PMID: 23165212
- PMCID: PMC3562302
- DOI: 10.4161/cc.22843
Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53
Abstract
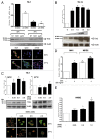
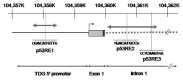
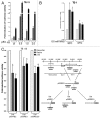
Thymine DNA glycosylase (TDG) belongs to the superfamily of uracil DNA glycosylases (UDG) and is the first enzyme in the base-excision repair pathway (BER) that removes thymine from G:T mismatches at CpG sites. This glycosylase activity has also been found to be critical for active demethylation of genes involved in embryonic development. Here we show that wild-type p53 transcriptionally regulates TDG expression. Chromatin immunoprecipitation (ChIP) and luciferase assays indicate that wild-type p53 binds to a domain of TDG promoter containing two p53 consensus response elements (p53RE) and activates its transcription. Next, we have used a panel of cell lines with different p53 status to demonstrate that TDG mRNA and protein expression levels are induced in a p53-dependent manner under different conditions. This panel includes isogenic breast and colorectal cancer cell lines with wild-type or inactive p53, esophageal squamous cell carcinoma cell lines lacking p53 or expressing a temperature-sensitive p53 mutant and normal human bronchial epithelial cells. Induction of TDG mRNA expression is accompanied by accumulation of TDG protein in both nucleus and cytoplasm, with nuclear re-localization occurring upon DNA damage in p53-competent, but not -incompetent, cells. These observations suggest a role for p53 activity in TDG nuclear translocation. Overall, our results show that TDG expression is directly regulated by p53, suggesting that loss of p53 function may affect processes mediated by TDG, thus negatively impacting on genetic and epigenetic stability.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
