Evolutionary conservation of Nkx2.5 autoregulation in the second heart field
- PMID: 23165293
- PMCID: PMC3549048
- DOI: 10.1016/j.ydbio.2012.11.007
Evolutionary conservation of Nkx2.5 autoregulation in the second heart field
Abstract
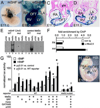
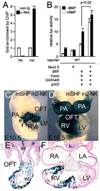
The cardiac homeobox gene Nkx2.5 plays a key and dosage-sensitive role in the differentiation of outflow tract and right ventricle from progenitors of the second heart field (SHF) and Nkx2.5 mutation is strongly associated with human outflow tract congenital heart disease (OFT CHD). Therefore defining the regulatory mechanisms controlling Nkx2.5 expression in SHF populations serves an important function in understanding the etiology of complex CHD. Through a comparative analysis of regulatory elements controlling SHF expression of Nkx2.5 in the chicken and mouse, we have found evidence that Nkx2.5 autoregulation is important for maintaining Nkx2.5 expression during SHF differentiation in both species. However the mechanism of Nkx2.5 maintenance differs between placental mammals and non-mammalian vertebrates: in chick Nkx2.5 binds directly to a genomic enhancer element that is required to maintain Nkx2.5 expression in the SHF. In addition, it is likely that this is true in other non-mammalian vertebrates given that they possess a similar genomic organization. By contrast, in placental mammals, Nkx2.5 autoregulation in the SHF functions indirectly through Mef2c. These data underscore a tight relationship in mammals between Nkx2.5 and Mef2c in SHF transcriptional regulation, and highlight the potential for evolutionary cis-regulatory analysis to identify core, conserved components of the gene networks controlling heart development.
Published by Elsevier Inc.
Figures




References
-
- Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development (Cambridge, England) 2002;129:1935–1943. - PubMed
-
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. - PubMed
-
- Barron M, Gao M, Lough J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev Dyn. 2000;218:383–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

