A family-based paradigm to identify candidate chromosomal regions for isolated congenital diaphragmatic hernia
- PMID: 23165927
- PMCID: PMC3507422
- DOI: 10.1002/ajmg.a.35664
A family-based paradigm to identify candidate chromosomal regions for isolated congenital diaphragmatic hernia
Abstract
Congenital diaphragmatic hernia (CDH) is a developmental defect of the diaphragm that causes high newborn mortality. Isolated or non-syndromic CDH is considered a multifactorial disease, with strong evidence implicating genetic factors. As low heritability has been reported in isolated CDH, family-based genetic methods have yet to identify the genetic factors associated with the defect. Using the Utah Population Database, we identified distantly related patients from several extended families with a high incidence of isolated CDH. Using high-density genotyping, seven patients were analyzed by homozygosity exclusion rare allele mapping (HERAM) and phased haplotype sharing (HapShare), two methods we developed to map shared chromosome regions. Our patient cohort shared three regions not previously associated with CDH, that is, 2q11.2-q12.1, 4p13 and 7q11.2, and two regions previously involved in CDH, that is, 8p23.1 and 15q26.2. The latter regions contain GATA4 and NR2F2, two genes implicated in diaphragm formation in mice. Interestingly, three patients shared the 8p23.1 locus and one of them also harbored the 15q26.2 segment. No coding variants were identified in GATA4 or NR2F2, but a rare shared variant was found in intron 1 of GATA4. This work shows the role of heritability in isolated CDH. Our family-based strategy uncovers new chromosomal regions possibly associated with disease, and suggests that non-coding variants of GATA4 and NR2F2 may contribute to the development of isolated CDH. This approach could speed up the discovery of the genes and regulatory elements causing multifactorial diseases, such as isolated CDH.
Copyright © 2012 Wiley Periodicals, Inc.
Figures
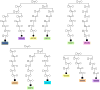






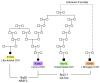
References
-
- Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am J Med Genet C Semin Med Genet. 2007;145C:109–116. - PubMed
-
- Arrington CB, Sower CT, Chuckwuk N, Stevens J, Leppert MF, Yetman AT, Bowles NE. Absence of TGFBR1 and TGFBR2 mutations in patients with bicuspid aortic valve and aortic dilation. Am J Cardiol. 2008;102:629–631. - PubMed
-
- Beurskens LW, Tibboel D, Lindemans J, Duvekot JJ, Cohen-Overbeek TE, Veenma DC, de Klein A, Greer JJ, Steegers-Theunissen RP. Retinol status of newborn infants is associated with congenital diaphragmatic hernia. Pediatrics. 2010;126:712–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

