Modulation of dendritic AMPA receptor mRNA trafficking by RNA splicing and editing
- PMID: 23166306
- PMCID: PMC3592400
- DOI: 10.1093/nar/gks1223
Modulation of dendritic AMPA receptor mRNA trafficking by RNA splicing and editing
Abstract
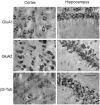
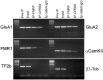
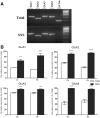
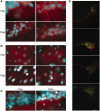
RNA trafficking to dendrites and local translation are crucial processes for superior neuronal functions. To date, several α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor (AMPAR) mRNAs have been detected in dendrites and are subject to local protein synthesis. Here, we report the presence of all AMPAR GluA1-4 mRNAs in hippocampal and cortical rat synaptic spines by synaptoneurosomes analysis. In particular, we showed that dendritic AMPAR mRNAs are present in the Flip versions in the cortex and hippocampus. To further confirm these data, we demonstrate, using in situ hybridization, the dendritic localization of the GluA2 Flip isoform in vitro and in vivo, whereas the Flop variant is restricted mainly to the soma. In addition, we report that dendritic AMPA mRNAs are edited at low levels at their R/G sites; this result was also supported with transfection experiments using chimeric GluA2 DNA vectors, showing that transcripts carrying an unedited nucleotide at the R/G site, in combination with the Flip exon, are more efficiently targeted to dendrites when compared with the edited-Flip versions. Our data show that post-transcriptional regulations such as RNA splicing, editing and trafficking might be mutually coordinated and that the localization of different AMPAR isoforms in dendrites might play a functional role in the regulation of neuronal transmission.
Figures


Similar articles
-
Cell class-specific regulation of neocortical dendrite and spine growth by AMPA receptor splice and editing variants.Development. 2011 Oct;138(19):4301-13. doi: 10.1242/dev.071076. Epub 2011 Aug 24. Development. 2011. PMID: 21865324
-
Differential dendritic targeting of AMPA receptor subunit mRNAs in adult rat hippocampal principal neurons and interneurons.J Comp Neurol. 2013 Jun 15;521(9):1954-2007. doi: 10.1002/cne.23292. J Comp Neurol. 2013. PMID: 23296627
-
Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons.J Neurosci. 2006 Aug 9;26(32):8339-51. doi: 10.1523/JNEUROSCI.0472-06.2006. J Neurosci. 2006. PMID: 16899729 Free PMC article.
-
The AMPAR subunit GluR2: still front and center-stage.Brain Res. 2000 Dec 15;886(1-2):190-207. doi: 10.1016/s0006-8993(00)02951-6. Brain Res. 2000. PMID: 11119696 Review.
-
Sculpting AMPA receptor formation and function by alternative RNA processing.RNA Biol. 2009 Nov-Dec;6(5):517-21. doi: 10.4161/rna.6.5.9552. Epub 2009 Nov 18. RNA Biol. 2009. PMID: 19717983 Review.
Cited by
-
The Good and the Bad of Glutamate Receptor RNA Editing.Mol Neurobiol. 2017 Nov;54(9):6795-6805. doi: 10.1007/s12035-016-0201-z. Epub 2016 Oct 20. Mol Neurobiol. 2017. PMID: 27766534 Review.
-
FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization.Nat Commun. 2015 May 13;6:7098. doi: 10.1038/ncomms8098. Nat Commun. 2015. PMID: 25968143 Free PMC article.
-
miR-9-5p is involved in the rescue of stress-dependent dendritic shortening of hippocampal pyramidal neurons induced by acute antidepressant treatment with ketamine.Neurobiol Stress. 2021 Aug 12;15:100381. doi: 10.1016/j.ynstr.2021.100381. eCollection 2021 Nov. Neurobiol Stress. 2021. PMID: 34458512 Free PMC article.
-
Cellular and genetic drivers of RNA editing variation in the human brain.Nat Commun. 2022 May 30;13(1):2997. doi: 10.1038/s41467-022-30531-0. Nat Commun. 2022. PMID: 35637184 Free PMC article.
-
Chronic mild stress induces anhedonic behavior and changes in glutamate release, BDNF trafficking and dendrite morphology only in stress vulnerable rats. The rapid restorative action of ketamine.Neurobiol Stress. 2019 Apr 2;10:100160. doi: 10.1016/j.ynstr.2019.100160. eCollection 2019 Feb. Neurobiol Stress. 2019. PMID: 31193464 Free PMC article.
References
-
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. - PubMed
-
- Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 2003;13:279–283. - PubMed
-
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. - PubMed
-
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. - PubMed

