Nuclear factor of activated T-cells (NFAT)C2 inhibits Notch receptor signaling in osteoblasts
- PMID: 23166323
- PMCID: PMC3537061
- DOI: 10.1074/jbc.M112.340455
Nuclear factor of activated T-cells (NFAT)C2 inhibits Notch receptor signaling in osteoblasts
Abstract
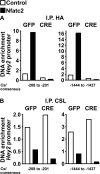
Notch receptors regulate osteoblastogenesis, and Notch activation induces cleavage and nuclear translocation of the Notch intracellular domain (NICD), which associates with Epstein-Barr virus latency C-promoter binding factor-1/suppressor of hairless/lag-1 (CSL) and induces transcription of Notch target genes, such as hairy enhancer of split-related with YRPW motif (Hey)1 and Hey2. Nuclear factors of activated T-cells (NFAT) are transcription factors that regulate osteoclastogenesis, but their function in osteoblasts is not clear. Notch inhibits NFATc1 transcription, but interactions between Notch and NFAT are understood poorly. To determine the regulation of NFAT expression by Notch, osteoblasts from Rosa(Notch) mice, where NICD is transcribed following excision of a loxP flanked STOP cassette, were used. Alternatively, wild-type C57BL/6 osteoblasts were exposed to the Notch ligand Delta-like (Dll)1 to induce Notch signaling or to bovine serum albumin as control. In Rosa(Notch) osteoblasts, Notch suppressed NFATc1 expression, increased Nfatc2 mRNA by post-transcriptional mechanisms, and had no effect on NFATc3 and NFATc4 transcripts. Induction of Nfatc2 transcripts by Notch was confirmed in C57BL/6 osteoblasts exposed to Dll1. To investigate NFATc2 function in osteoblasts, constitutively active NFATc2 was overexpressed in Rosa(Notch) osteoblasts. NFATc2 suppressed Notch transactivation and expression of Hey genes. Electrophoretic mobility shift assays revealed that NFATc2 and CSL bind to similar DNA sequences, and chromatin immunoprecipitation indicated that NFATc2 displaced CSL from the Hey2 promoter. The effects of NICD and NFATc2 in Rosa(Notch) osteoblasts were assessed, and both proteins inhibited osteoblast function. In conclusion, Notch stabilizes Nfatc2 transcripts, NFATc2 suppresses Notch signaling, and both proteins inhibit osteoblast function.
Figures





Similar articles
-
Reciprocal regulation of Notch and nuclear factor of activated T-cells (NFAT) c1 transactivation in osteoblasts.J Biol Chem. 2011 Feb 11;286(6):4576-88. doi: 10.1074/jbc.M110.161893. Epub 2010 Dec 3. J Biol Chem. 2011. PMID: 21131365 Free PMC article.
-
Notch suppresses nuclear factor of activated T cells (NFAT) transactivation and Nfatc1 expression in chondrocytes.Endocrinology. 2013 Feb;154(2):762-72. doi: 10.1210/en.2012-1925. Epub 2012 Dec 21. Endocrinology. 2013. PMID: 23264614 Free PMC article.
-
Glucocorticoids inhibit notch target gene expression in osteoblasts.J Cell Biochem. 2018 Jul;119(7):6016-6023. doi: 10.1002/jcb.26798. Epub 2018 Mar 25. J Cell Biochem. 2018. PMID: 29575203 Free PMC article.
-
Notch and the skeleton.Mol Cell Biol. 2010 Feb;30(4):886-96. doi: 10.1128/MCB.01285-09. Epub 2009 Dec 7. Mol Cell Biol. 2010. PMID: 19995916 Free PMC article. Review.
-
Notch Signaling and the Skeleton.Endocr Rev. 2016 Jun;37(3):223-53. doi: 10.1210/er.2016-1002. Epub 2016 Apr 13. Endocr Rev. 2016. PMID: 27074349 Free PMC article. Review.
Cited by
-
Hajdu Cheney Mouse Mutants Exhibit Osteopenia, Increased Osteoclastogenesis, and Bone Resorption.J Biol Chem. 2016 Jan 22;291(4):1538-1551. doi: 10.1074/jbc.M115.685453. Epub 2015 Dec 1. J Biol Chem. 2016. PMID: 26627824 Free PMC article.
-
Hairy and Enhancer of Split-Related With YRPW Motif-Like (HeyL) Is Dispensable for Bone Remodeling in Mice.J Cell Biochem. 2017 Jul;118(7):1819-1826. doi: 10.1002/jcb.25859. Epub 2017 Mar 9. J Cell Biochem. 2017. PMID: 28019674 Free PMC article.
-
NFAT signaling in osteoblasts regulates the hematopoietic niche in the bone microenvironment.Clin Dev Immunol. 2013;2013:107321. doi: 10.1155/2013/107321. Epub 2013 Sep 1. Clin Dev Immunol. 2013. PMID: 24023563 Free PMC article.
-
The Hajdu Cheney mutation sensitizes mice to the osteolytic actions of tumor necrosis factor α.J Biol Chem. 2019 Sep 27;294(39):14203-14214. doi: 10.1074/jbc.RA119.009824. Epub 2019 Aug 1. J Biol Chem. 2019. PMID: 31371452 Free PMC article.
-
Dynamic transcriptome analysis of NFAT family in guided bone regeneration with occlusive periosteum in swine model.J Orthop Surg Res. 2022 Jul 26;17(1):364. doi: 10.1186/s13018-022-03252-9. J Orthop Surg Res. 2022. PMID: 35883195 Free PMC article.
References
-
- Fortini M. E. (2009) Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 - PubMed
-
- Ehebauer M., Hayward P., Martinez-Arias A. (2006) Notch signaling pathway. Sci. STKE 2006, cm7. - PubMed
-
- Kovall R. A. (2008) More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27, 5099–5109 - PubMed
-
- Iso T., Kedes L., Hamamori Y. (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194, 237–255 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

