Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics
- PMID: 23166327
- PMCID: PMC3537059
- DOI: 10.1074/jbc.M112.425736
Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics
Abstract
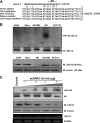
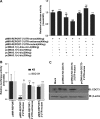
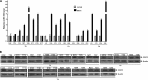
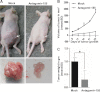
The CDC73 gene is mutationally inactivated in hereditary and sporadic parathyroid tumors. It negatively regulates β-catenin, cyclin D1, and c-MYC. Down-regulation of CDC73 has been reported in breast, renal, and gastric carcinomas. However, the reports regarding the role of CDC73 in oral squamous cell carcinoma (OSCC) are lacking. In this study we show that CDC73 is down-regulated in a majority of OSCC samples. We further show that oncogenic microRNA-155 (miR-155) negatively regulates CDC73 expression. Our experiments show that the dramatic up-regulation of miR-155 is an exclusive mechanism for down-regulation of CDC73 in a panel of human cell lines and a subset of OSCC patient samples in the absence of loss of heterozygosity, mutations, and promoter methylation. Ectopic expression of miR-155 in HEK293 cells dramatically reduced CDC73 levels, enhanced cell viability, and decreased apoptosis. Conversely, the delivery of a miR-155 antagonist (antagomir-155) to KB cells overexpressing miR-155 resulted in increased CDC73 levels, decreased cell viability, increased apoptosis, and marked regression of xenografts in nude mice. Cotransfection of miR-155 with CDC73 in HEK293 cells abrogated its pro-oncogenic effect. Reduced cell proliferation and increased apoptosis of KB cells were dependent on the presence or absence of the 3'-UTR in CDC73. In summary, knockdown of CDC73 expression due to overexpression of miR-155 not only adds a novelty to the list of mechanisms responsible for its down-regulation in different tumors, but the restoration of CDC73 levels by the use of antagomir-155 may also have an important role in therapeutic intervention of cancers, including OSCC.
Figures

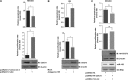
References
-
- Shattuck T. M., Välimäki S., Obara T., Gaz R. D., Clark O.H., Shoback D., Wierman M. E., Tojo K., Robbins C. M., Carpten J. D., Farnebo L. O., Larsson C., Arnold A. (2003) Somatic and germ line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N. Engl. J. Med. 349, 1722–1729 - PubMed
-
- Aldred M. J., Talacko A. A., Savarirayan R., Murdolo V., Mills A. E., Radden B. G., Alimov A., Villablanca A., Larsson C. (2006) Dental findings in a family with hyperparathyroidism. Jaw tumour syndrome and a novel HRPT2 gene mutation. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 101, 212–218 - PubMed
-
- Pimenta F. J., Gontijo Silveira L.F., Tavares G. C., Silva A. C., Perdigão P. F., Castro W. H., Gomez M. V., Teh B. T., De Marco L., Gomez R. S. (2006) HRPT2 gene alterations in ossifying fibroma of the jaws. Oral. Oncol. 42, 735–739 - PubMed
-
- Carpten J. D., Robbins C. M., Villablanca A., Forsberg L., Presciuttini S., Bailey-Wilson J., Simonds W. F., Gillanders E. M., Kennedy A. M., Chen J. D., Agarwal S. K., Sood R., Jones M. P., Moses T. Y., Haven C., Petillo D., Leotlela P. D., Harding B., Cameron D., Pannett A. A., Höög A., Heath H., 3rd, James-Newton L. A., Robinson B., Zarbo R. J., Cavaco B. M., Wassif W., Perrier N. D., Rosen I. B., Kristoffersson U., Turnpenny P. D., Farnebo L. O., Besser G. M., Jackson C. E., Morreau H., Trent J. M., Thakker R. V., Marx S. J., Teh B. T., Larsson C., Hobbs M. R. (2002) HRPT2, encoding parafibromin, is mutated in hyperparathyroidism. Jaw tumour syndrome. Nat. Genet. 32, 676–680 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

