DNA methylation dynamics during the mammalian life cycle
- PMID: 23166392
- PMCID: PMC3539357
- DOI: 10.1098/rstb.2011.0328
DNA methylation dynamics during the mammalian life cycle
Abstract
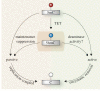
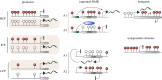
DNA methylation is dynamically remodelled during the mammalian life cycle through distinct phases of reprogramming and de novo methylation. These events enable the acquisition of cellular potential followed by the maintenance of lineage-restricted cell identity, respectively, a process that defines the life cycle through successive generations. DNA methylation contributes to the epigenetic regulation of many key developmental processes including genomic imprinting, X-inactivation, genome stability and gene regulation. Emerging sequencing technologies have led to recent insights into the dynamic distribution of DNA methylation during development and the role of this epigenetic mark within distinct genomic contexts, such as at promoters, exons or imprinted control regions. Additionally, there is a better understanding of the mechanistic basis of DNA demethylation during epigenetic reprogramming in primordial germ cells and during pre-implantation development. Here, we discuss our current understanding of the developmental roles and dynamics of this key epigenetic system.
Figures

References
-
- Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–43210.1038/nature05918 (doi:10.1038/nature05918) - DOI - DOI - PubMed
-
- Surani MA, Hayashi K, Hajkova P. 2007. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–76210.1016/j.cell.2007.02.010 (doi:10.1016/j.cell.2007.02.010) - DOI - DOI - PubMed
-
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–2110.1101/gad.947102 (doi:10.1101/gad.947102) - DOI - DOI - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–25710.1016/S0092-8674(00)81656-6 (doi:10.1016/S0092-8674(00)81656-6) - DOI - DOI - PubMed
-
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–92610.1016/0092-8674(92)90611-F (doi:10.1016/0092-8674(92)90611-F) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
