The origin and evolution of genomic imprinting and viviparity in mammals
- PMID: 23166401
- PMCID: PMC3539366
- DOI: 10.1098/rstb.2012.0151
The origin and evolution of genomic imprinting and viviparity in mammals
Abstract
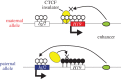
Genomic imprinting is widespread in eutherian mammals. Marsupial mammals also have genomic imprinting, but in fewer loci. It has long been thought that genomic imprinting is somehow related to placentation and/or viviparity in mammals, although neither is restricted to mammals. Most imprinted genes are expressed in the placenta. There is no evidence for genomic imprinting in the egg-laying monotreme mammals, despite their short-lived placenta that transfers nutrients from mother to embryo. Post natal genomic imprinting also occurs, especially in the brain. However, little attention has been paid to the primary source of nutrition in the neonate in all mammals, the mammary gland. Differentially methylated regions (DMRs) play an important role as imprinting control centres in each imprinted region which usually comprises both paternally and maternally expressed genes (PEGs and MEGs). The DMR is established in the male or female germline (the gDMR). Comprehensive comparative genome studies demonstrated that two imprinted regions, PEG10 and IGF2-H19, are conserved in both marsupials and eutherians and that PEG10 and H19 DMRs emerged in the therian ancestor at least 160 Ma, indicating the ancestral origin of genomic imprinting during therian mammal evolution. Importantly, these regions are known to be deeply involved in placental and embryonic growth. It appears that most maternal gDMRs are always associated with imprinting in eutherian mammals, but emerged at differing times during mammalian evolution. Thus, genomic imprinting could evolve from a defence mechanism against transposable elements that depended on DNA methylation established in germ cells.
Figures


References
-
- Luo ZX, Yuan CX, Meng QJ, Ji Q. 2011. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476, 442–445 10.1038/nature10291 (doi:10.1038/nature10291) - DOI - PubMed
-
- Kaneko-Ishino T, Ishino F. 2010. Retrotransposon silencing by DNA methylation contributed to the evolution of placentation and genomic imprinting in mammals. Dev. Growth Differ. 52, 533–543 10.1111/j.1440-169X.2010.01194.x (doi:10.1111/j.1440-169X.2010.01194.x) - DOI - PubMed
-
- Mossman HW. 1987. Vertebrate fetal membranes. New Brunswick, NJ: Rutgers University Press
-
- Amoroso EC, Heap RB, Renfree MB. 1979. Hormones and the evolution of viviparity. In Hormones and evolution (ed. Barrington JW.), pp. 925–989 New York, NY: Academic Press
-
- Renfree MB. 1982. Implantation and placentation. In Reproduction in mammals (eds Austin CR, Short RV.), pp. 26–69, 2nd edn Cambridge, UK: Cambridge University Press
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
