This Déjà vu feeling--analysis of multidomain protein evolution in eukaryotic genomes
- PMID: 23166479
- PMCID: PMC3499355
- DOI: 10.1371/journal.pcbi.1002701
This Déjà vu feeling--analysis of multidomain protein evolution in eukaryotic genomes
Abstract
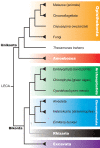
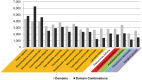
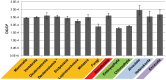
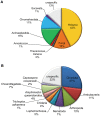
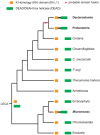
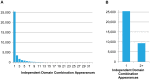
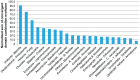
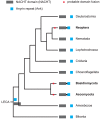
Evolutionary innovation in eukaryotes and especially animals is at least partially driven by genome rearrangements and the resulting emergence of proteins with new domain combinations, and thus potentially novel functionality. Given the random nature of such rearrangements, one could expect that proteins with particularly useful multidomain combinations may have been rediscovered multiple times by parallel evolution. However, existing reports suggest a minimal role of this phenomenon in the overall evolution of eukaryotic proteomes. We assembled a collection of 172 complete eukaryotic genomes that is not only the largest, but also the most phylogenetically complete set of genomes analyzed so far. By employing a maximum parsimony approach to compare repertoires of Pfam domains and their combinations, we show that independent evolution of domain combinations is significantly more prevalent than previously thought. Our results indicate that about 25% of all currently observed domain combinations have evolved multiple times. Interestingly, this percentage is even higher for sets of domain combinations in individual species, with, for instance, 70% of the domain combinations found in the human genome having evolved independently at least once in other species. We also show that previous, much lower estimates of this rate are most likely due to the small number and biased phylogenetic distribution of the genomes analyzed. The process of independent emergence of identical domain combination is widespread, not limited to domains with specific functional categories. Besides data from large-scale analyses, we also present individual examples of independent domain combination evolution. The surprisingly large contribution of parallel evolution to the development of the domain combination repertoire in extant genomes has profound consequences for our understanding of the evolution of pathways and cellular processes in eukaryotes and for comparative functional genomics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Evolutionary versatility of eukaryotic protein domains revealed by their bigram networks.BMC Evol Biol. 2011 Aug 18;11:242. doi: 10.1186/1471-2148-11-242. BMC Evol Biol. 2011. PMID: 21849086 Free PMC article.
-
The modular nature of protein evolution: domain rearrangement rates across eukaryotic life.BMC Evol Biol. 2020 Feb 14;20(1):30. doi: 10.1186/s12862-020-1591-0. BMC Evol Biol. 2020. PMID: 32059645 Free PMC article.
-
Function-selective domain architecture plasticity potentials in eukaryotic genome evolution.Biochimie. 2015 Dec;119:269-77. doi: 10.1016/j.biochi.2015.05.003. Epub 2015 May 15. Biochimie. 2015. PMID: 25980317 Free PMC article.
-
Origin and evolution of eukaryotic transcription factors.Curr Opin Genet Dev. 2019 Oct;58-59:25-32. doi: 10.1016/j.gde.2019.07.010. Epub 2019 Aug 26. Curr Opin Genet Dev. 2019. PMID: 31466037 Review.
-
Genome and protein evolution in eukaryotes.Curr Opin Chem Biol. 2002 Feb;6(1):39-45. doi: 10.1016/s1367-5931(01)00278-2. Curr Opin Chem Biol. 2002. PMID: 11827821 Review.
Cited by
-
Analysis of the protein domain and domain architecture content in fungi and its application in the search of new antifungal targets.PLoS Comput Biol. 2014 Jul 17;10(7):e1003733. doi: 10.1371/journal.pcbi.1003733. eCollection 2014 Jul. PLoS Comput Biol. 2014. PMID: 25033262 Free PMC article.
-
Plant Proteins Are Smaller Because They Are Encoded by Fewer Exons than Animal Proteins.Genomics Proteomics Bioinformatics. 2016 Dec;14(6):357-370. doi: 10.1016/j.gpb.2016.06.003. Epub 2016 Dec 18. Genomics Proteomics Bioinformatics. 2016. PMID: 27998811 Free PMC article.
-
Dynamics of genomic innovation in the unicellular ancestry of animals.Elife. 2017 Jul 20;6:e26036. doi: 10.7554/eLife.26036. Elife. 2017. PMID: 28726632 Free PMC article.
-
AIDA: ab initio domain assembly for automated multi-domain protein structure prediction and domain-domain interaction prediction.Bioinformatics. 2015 Jul 1;31(13):2098-105. doi: 10.1093/bioinformatics/btv092. Epub 2015 Feb 19. Bioinformatics. 2015. PMID: 25701568 Free PMC article.
-
Similar Ratios of Introns to Intergenic Sequence across Animal Genomes.Genome Biol Evol. 2017 Jun 1;9(6):1582-1598. doi: 10.1093/gbe/evx103. Genome Biol Evol. 2017. PMID: 28633296 Free PMC article.
References
-
- Moore AD, Björklund ÅK, Ekman D, Bornberg-Bauer E, Elofsson A (2008) Arrangements in the modular evolution of proteins. Trends Biochem Sci 33: 444–451. - PubMed
-
- Jin J, Xie X, Chen C, Park JG, Stark C, et al. (2009) Eukaryotic protein domains as functional units of cellular evolution. Sci Signal 2: ra76. - PubMed

